Workflow Type: Galaxy
Frozen
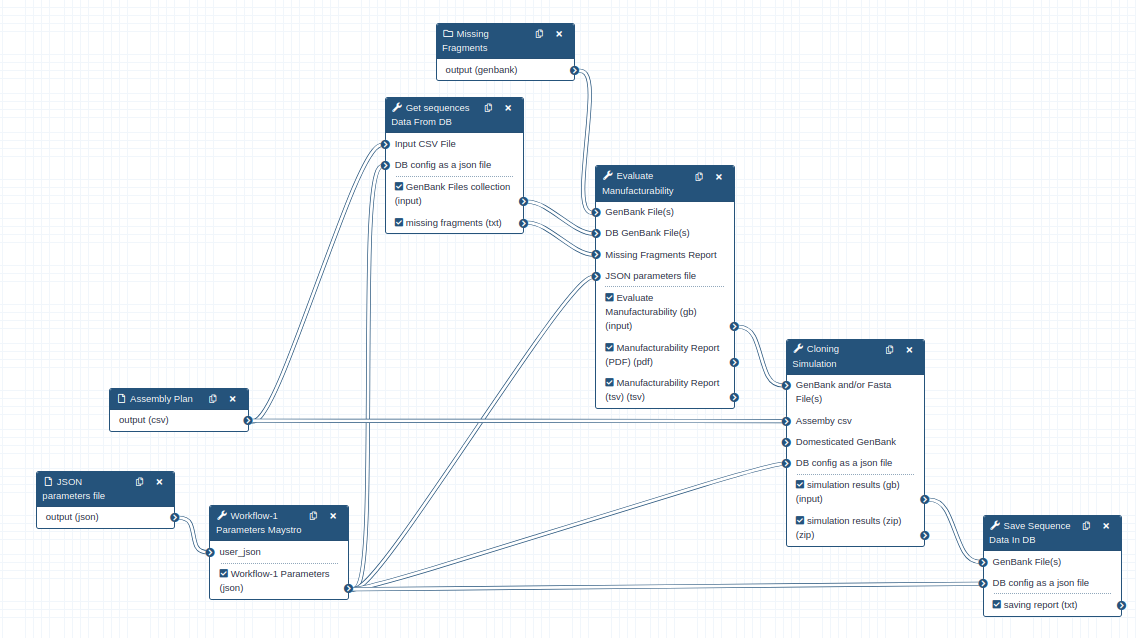
Cloning simulation workflow for sequences present in DB
Run the GoldenGate cloning simulation for a list of constructs in a CSV file and interact with a database
inputs:
- Assmbly Plan (without header) : The CSV file should contain the constraints line by line in the first column, along with their associated fragments on each line. This data will be passed to the seq_from_DB tool.
- JSON parameters file (optional) : The JSON file should contain the the parameters used in workflow tools. If this input is not set, user should set the parameters mannualy in the maystro tool.
- Missing Fragments (optional) : The collection of GenBank files that are not present in the database ( make collection). If a fragment is listed in the Assembly Plan CSV but missing from the database, it must be provided through this input.
steps:
workflow_1 Parameter_Maystro
- Distribute workflow parameters on the workflow tools
- Parameters can be set as a JSON file in input (optional)
- Parameters can be manually instate of JSON file
- For the first use, it is recommended to set the parameters manually in the tool. A JSON file will then be generated automatically for use in subsequent runs.
seq_from_DB
- Extract the fragments associated with each constraint from the CSV file.
- Check if all fragments are present in the database.
evaluate_manufacturability
- If any fragment is missing it will serve from the unannotated Genbenk provided to annotate them and add them to the finale gb collection
- If all fragments are present in the database, GenBank files for each fragment will be passed to the cloning_simulation tool.
clonning_simulation
- GoldenGate cloning simulation based on constraints.
- Generate GenBank files for each simulated constraint.
seq_to_db
- Constraint GenBank files can be saved to the database, depending on the user's choice.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Assembly Plan | #main/Assembly Plan | n/a |
|
| JSON parameters file | #main/JSON parameters file | n/a |
|
| Missing Fragments | #main/Missing Fragments | GenBank collection for fragments not found in the database |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Workflow-1 Parameters Maystro | toolshed.g2.bx.psu.edu/repos/tduigou/parameters_maystro_workflow_1/parameters_maystro_workflow_1/0.1.0 |
| 4 | Get sequences Data From DB | toolshed.g2.bx.psu.edu/repos/tduigou/seq_from_db/seq_form_db/0.3.0+galaxy2 |
| 5 | Evaluate Manufacturability | toolshed.g2.bx.psu.edu/repos/tduigou/evaluate_manufacturability/evaluate_manufacturability/0.3.0+galaxy2 |
| 6 | Cloning Simulation | toolshed.g2.bx.psu.edu/repos/tduigou/cloning_simulation/cloning_simulation/0.2.0+galaxy1 |
| 7 | Save Sequence Data In DB | toolshed.g2.bx.psu.edu/repos/tduigou/seq_to_db/seq_to_db/0.3.0+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Evaluate Manufacturability (gb) | #main/Evaluate Manufacturability (gb) | n/a |
|
| GenBank Files collection | #main/GenBank Files collection | n/a |
|
| Manufacturability Report (PDF) | #main/Manufacturability Report (PDF) | n/a |
|
| Manufacturability Report (tsv) | #main/Manufacturability Report (tsv) | n/a |
|
| Workflow-1 Parameters | #main/Workflow-1 Parameters | n/a |
|
| missing fragments | #main/missing fragments | n/a |
|
| saving report | #main/saving report | n/a |
|
| simulation results (gb) | #main/simulation results (gb) | n/a |
|
| simulation results (zip) | #main/simulation results (zip) | n/a |
|
Version History
Version 2 (earliest) Created 17th Oct 2025 at 12:22 by ramiz khaled
workflow 0 + 1
Frozen
 Version-2
Version-27daa0d1
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Activity
Views: 412 Downloads: 69 Runs: 1
Created: 17th Oct 2025 at 12:22
Last updated: 17th Oct 2025 at 12:41
Annotated Properties
Topic annotations
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy