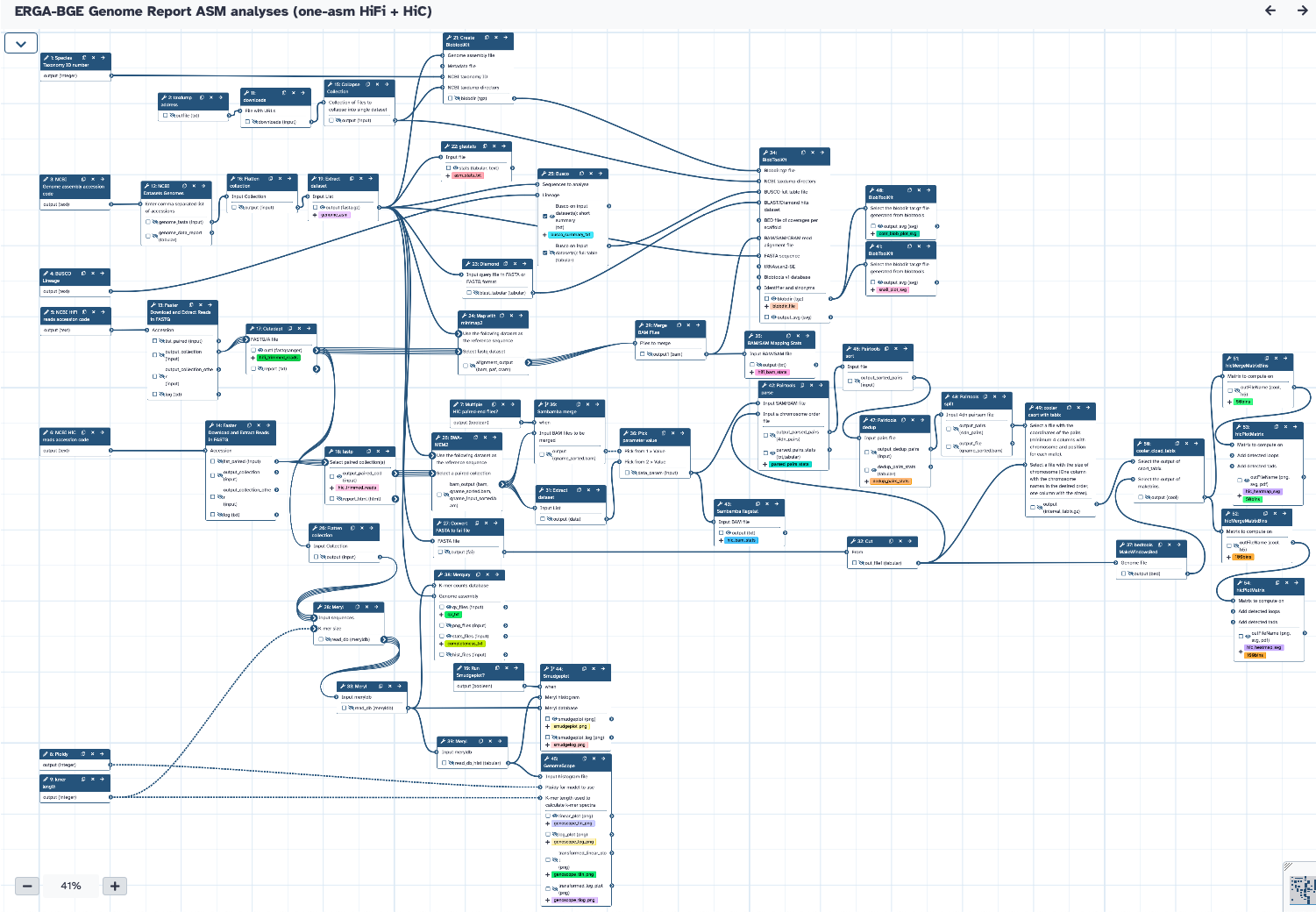
Workflow Type: Galaxy
Open
Frozen
Stable
Assembly Evaluation for ERGA-BGE Reports
One Assembly, HiFi WGS reads + HiC reads
The workflow requires the following:
- Species Taxonomy ID number
- NCBI Genome assembly accession code
- BUSCO Lineage
- WGS accurate reads accession code
- NCBI HiC reads accession code
The workflow will get the data and process it to generate genome profiling (genomescope, smudgeplot -optional-), assembly stats (gfastats), merqury stats (QV, completeness), BUSCO, snailplot, contamination blobplot, and HiC heatmap.
Use this workflow for HiFi-based assemblies where the WGS accurate reads are PacBio HiFi
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| BUSCO Lineage | BUSCO Lineage | Choose the (eukaryotic) BUSCO lineage that corresponds to the assembled species, e.g.: mammalia_odb10 |
|
| Multiple HiC paired-end files? | Multiple HiC paired-end files? | IMPORTANT! If you entered more than one accession code, select Yes |
|
| NCBI Genome assembly accession code | NCBI Genome assembly accession code | Should start with GCA or GCF, e.g.: GCA_963556495.2 |
|
| NCBI HiC reads accession code | NCBI HiC reads accession code | Comma-separated accession code of the reads. Must start with SRR, DRR or ERR, e.g. SRR925743, ERR343809 |
|
| NCBI HiFi reads accession code | NCBI HiFi reads accession code | Comma-separated accession code of the reads. Must start with SRR, DRR or ERR, e.g. SRR925743, ERR343809 |
|
| Ploidy | Ploidy | Default value: 2 |
|
| Run Smudgeplot? | Run Smudgeplot? | n/a |
|
| Species Taxonomy ID number | Species Taxonomy ID number | Get the NCBI taxonomy number here: https://www.ncbi.nlm.nih.gov/taxonomy |
|
| kmer length | kmer length | Default value: 21 |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | taxdump address | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_text_file_with_recurring_lines/9.3+galaxy1 |
| 10 | downloads | lftp |
| 11 | NCBI Datasets Genomes | toolshed.g2.bx.psu.edu/repos/iuc/ncbi_datasets/datasets_download_genome/16.20.0+galaxy0 |
| 12 | Faster Download and Extract Reads in FASTQ | toolshed.g2.bx.psu.edu/repos/iuc/sra_tools/fasterq_dump/3.1.1+galaxy0 |
| 13 | Faster Download and Extract Reads in FASTQ | toolshed.g2.bx.psu.edu/repos/iuc/sra_tools/fasterq_dump/3.1.1+galaxy0 |
| 14 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 15 | Flatten collection | __FLATTEN__ |
| 16 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/4.9+galaxy1 |
| 17 | fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.23.4+galaxy0 |
| 18 | Extract dataset | __EXTRACT_DATASET__ |
| 19 | Flatten collection | __FLATTEN__ |
| 20 | Create BlobtoolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 21 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.6+galaxy0 |
| 22 | Diamond | toolshed.g2.bx.psu.edu/repos/bgruening/diamond/bg_diamond/2.0.15+galaxy0 |
| 23 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy0 |
| 24 | Busco | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.5.0+galaxy0 |
| 25 | BWA-MEM2 | toolshed.g2.bx.psu.edu/repos/iuc/bwa_mem2/bwa_mem2/2.2.1+galaxy1 |
| 26 | Convert FASTA to fai file | CONVERTER_fasta_to_fai |
| 27 | Meryl | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 28 | Merge BAM Files | toolshed.g2.bx.psu.edu/repos/devteam/sam_merge/sam_merge2/1.2.0 |
| 29 | Sambamba merge | toolshed.g2.bx.psu.edu/repos/bgruening/sambamba_merge/sambamba_merge/1.0.1+galaxy1 |
| 30 | Extract dataset | __EXTRACT_DATASET__ |
| 31 | Cut | Cut1 |
| 32 | Meryl | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 33 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 34 | BAM/SAM Mapping Stats | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_bam_stat/5.0.3+galaxy0 |
| 35 | Pick parameter value | toolshed.g2.bx.psu.edu/repos/iuc/pick_value/pick_value/0.2.0 |
| 36 | bedtools MakeWindowsBed | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_makewindowsbed/2.31.1 |
| 37 | Merqury | toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy3 |
| 38 | Meryl | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 39 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 40 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 41 | Pairtools parse | toolshed.g2.bx.psu.edu/repos/iuc/pairtools_parse/pairtools_parse/1.1.0+galaxy1 |
| 42 | Sambamba flagstat | toolshed.g2.bx.psu.edu/repos/bgruening/sambamba_flagstat/sambamba_flagstat/1.0.1+galaxy1 |
| 43 | Smudgeplot | toolshed.g2.bx.psu.edu/repos/galaxy-australia/smudgeplot/smudgeplot/0.2.5+galaxy3 |
| 44 | GenomeScope | toolshed.g2.bx.psu.edu/repos/iuc/genomescope/genomescope/2.0+galaxy2 |
| 45 | Pairtools sort | toolshed.g2.bx.psu.edu/repos/iuc/pairtools_sort/pairtools_sort/1.1.0+galaxy1 |
| 46 | Pairtools dedup | toolshed.g2.bx.psu.edu/repos/iuc/pairtools_dedup/pairtools_dedup/1.1.0+galaxy1 |
| 47 | Pairtools split | toolshed.g2.bx.psu.edu/repos/iuc/pairtools_split/pairtools_split/1.1.0+galaxy1 |
| 48 | cooler csort with tabix | toolshed.g2.bx.psu.edu/repos/lldelisle/cooler_csort_tabix/cooler_csort_tabix/0.8.11+galaxy1 |
| 49 | cooler_cload_tabix | toolshed.g2.bx.psu.edu/repos/lldelisle/cooler_cload_tabix/cooler_cload_tabix/0.8.11+galaxy1 |
| 50 | hicMergeMatrixBins | toolshed.g2.bx.psu.edu/repos/bgruening/hicexplorer_hicmergematrixbins/hicexplorer_hicmergematrixbins/3.7.2+galaxy0 |
| 51 | hicMergeMatrixBins | toolshed.g2.bx.psu.edu/repos/bgruening/hicexplorer_hicmergematrixbins/hicexplorer_hicmergematrixbins/3.7.2+galaxy0 |
| 52 | hicPlotMatrix | toolshed.g2.bx.psu.edu/repos/bgruening/hicexplorer_hicplotmatrix/hicexplorer_hicplotmatrix/3.7.2+galaxy0 |
| 53 | hicPlotMatrix | toolshed.g2.bx.psu.edu/repos/bgruening/hicexplorer_hicplotmatrix/hicexplorer_hicplotmatrix/3.7.2+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Busco on input dataset(s): short summary | Busco on input dataset(s): short summary | n/a |
|
| Busco on input dataset(s): full table | Busco on input dataset(s): full table | n/a |
|
Version History
Version 1.1 (latest) Created 4th Nov 2024 at 14:29 by Diego De Panis
No revision comments
Open
 master
master9b0d0d4
Version 1 (earliest) Created 20th Aug 2024 at 14:19 by Diego De Panis
Initial commit
Frozen
 Version-1
Version-148bc4d9
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
ERGA
Submitter
Discussion Channel
Tools
License
Activity
Views: 4275 Downloads: 738 Runs: 56
Created: 20th Aug 2024 at 14:19
Last updated: 5th Dec 2024 at 16:47
Annotated Properties
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-3679-9585
https://orcid.org/0000-0002-3679-9585