Workflow Type: Galaxy
Frozen
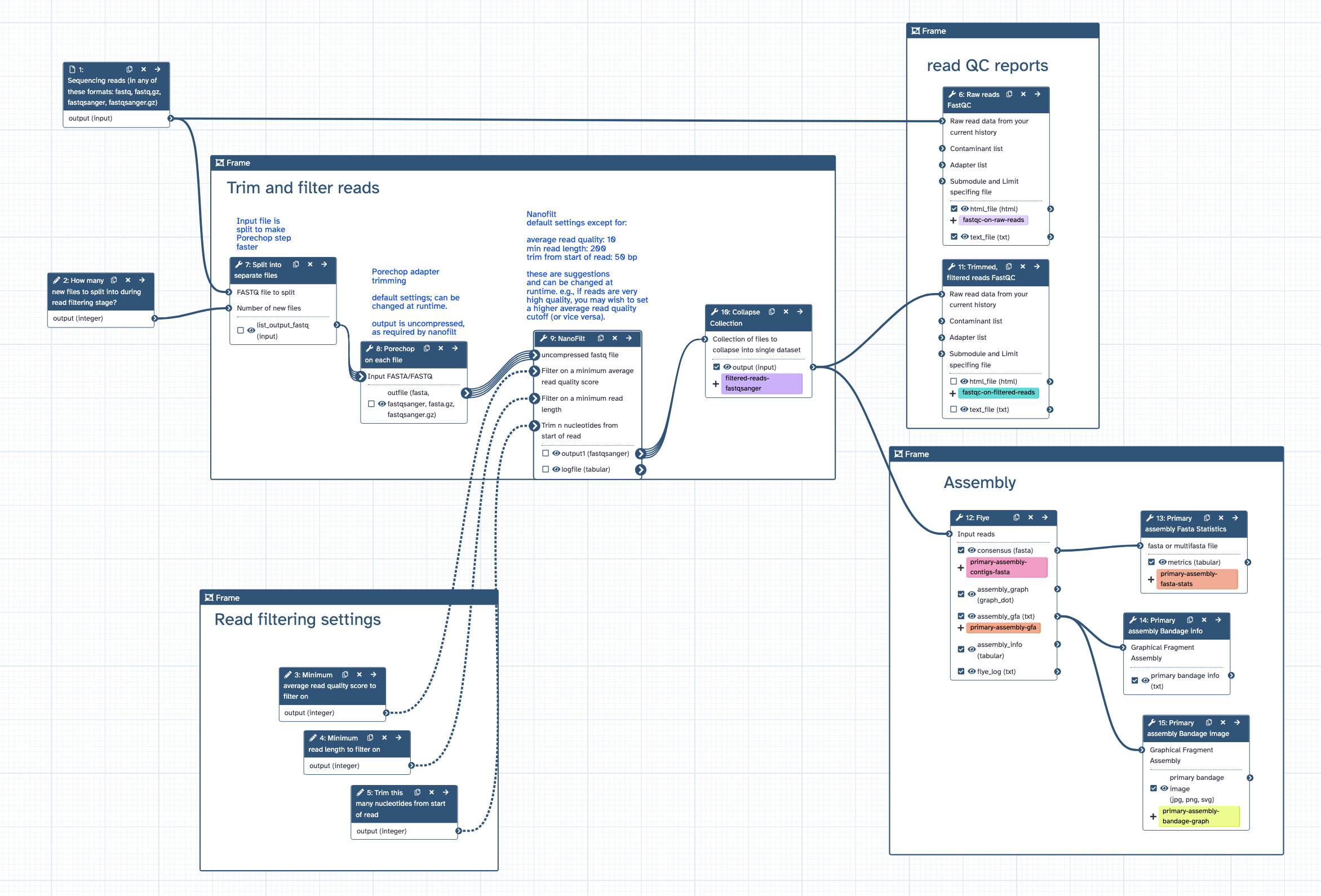
Genome assembly workflow for nanopore reads, for TSI
Input:
- Nanopore reads (can be in format: fastq, fastq.gz, fastqsanger, or fastqsanger.gz)
Optional settings to specify when the workflow is run:
- [1] how many input files to split the original input into (to speed up the workflow). default = 0. example: set to 2000 to split a 60 GB read file into 2000 files of ~ 30 MB.
- [2] filtering: min average read quality score. default = 10
- [3] filtering: min read length. default = 200
- [4] trimming: trim this many nucleotides from start of read. default = 50
- [5] note: these are suggestions and will depend on the characteristics of your raw reads and downstream aims. If filtering and trimming settings are too stringent, there may be no reads remaining and workflow will fail.
Workflow steps:
- [1] runs FastQC on raw reads
- [2] splits input reads file into separate files to speed up the next step of Porechop
- [3] trims nanopore adapters using Porechop
- [4] trims and filters nanopore reads by quality and length using Nanofilt
- [5] collapses back into a single read file, fastqsanger format
- [6] runs FastqQC on trimmed/filtered reads
- [7] assembles genome with Flye
- [8] calculates statistics on genome assembly contigs with Fasta Statistics
- [9] draws genome assembly graph with Bandage
Main outputs:
- [1] FastQC report on raw reads, html
- [2] Adpater-chopped, trimmed, filtered reads in fastqsanger format
- [3] FastQC report on filtered reads, html
- [4] genome assembly contigs in fasta format (primary assembly)
- [5] genome assembly statistics
- [6] genome assembly graph in Bandage format
Note: You may wish to plot raw reads first (e.g. using the tool NanoPlot), to get a better of idea of read lengths and quality, to decide on filtering/trimming settings.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| How many new files to split into during read filtering stage? | How many new files to split into during read filtering stage? | Split input to speed up next step with Porechop. e.g. if input fastq is 60 GB, split into 2000 files of approx 30 MB. |
|
| Minimum average read quality score to filter on | Minimum average read quality score to filter on | n/a |
|
| Minimum read length to filter on | Minimum read length to filter on | n/a |
|
| Sequencing reads (in any of these formats: fastq, fastq.gz, fastqsanger, fastqsanger.gz) | Sequencing reads (in any of these formats: fastq, fastq.gz, fastqsanger, fastqsanger.gz) | n/a |
|
| Trim this many nucleotides from start of read | Trim this many nucleotides from start of read | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 5 | Raw reads FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 6 | Split into separate files | toolshed.g2.bx.psu.edu/repos/bgruening/split_file_to_collection/split_file_to_collection/0.5.2 |
| 7 | Porechop on each file | toolshed.g2.bx.psu.edu/repos/iuc/porechop/porechop/0.2.4+galaxy0 |
| 8 | NanoFilt | toolshed.g2.bx.psu.edu/repos/leomrtns/nanofilt/nanofilt/0.1.0 |
| 9 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 10 | Trimmed, filtered reads FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 11 | Flye | default setting changed: remove non-primary contigs from assembly = yes toolshed.g2.bx.psu.edu/repos/bgruening/flye/flye/2.9.3+galaxy0 |
| 12 | Primary assembly Fasta Statistics | toolshed.g2.bx.psu.edu/repos/iuc/fasta_stats/fasta-stats/1.0.3 |
| 13 | Primary assembly Bandage info | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_info/0.8.1+galaxy1 |
| 14 | Primary assembly Bandage image | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/0.8.1+galaxy3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| text_file | text_file | n/a |
|
| html_file | html_file | n/a |
|
| output | output | n/a |
|
| assembly_graph | assembly_graph | n/a |
|
| assembly_gfa | assembly_gfa | n/a |
|
| assembly_info | assembly_info | n/a |
|
| flye_log | flye_log | n/a |
|
| consensus | consensus | n/a |
|
| metrics | metrics | n/a |
|
| primary bandage info | primary bandage info | n/a |
|
| primary bandage image | primary bandage image | n/a |
|
Version History
Version 1 (earliest) Created 3rd Sep 2024 at 02:07 by Anna Syme
Initial commit
Frozen
 Version-1
Version-1fc40931
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Syme, A. (2024). Genome assembly workflow for nanopore reads, for TSI. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.1114.1
Activity
Views: 3589 Downloads: 435 Runs: 3
Created: 3rd Sep 2024 at 02:07
Last updated: 3rd Sep 2024 at 02:13
 Tags
Tags Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673