Workflow Type: Python
Frozen
Stable
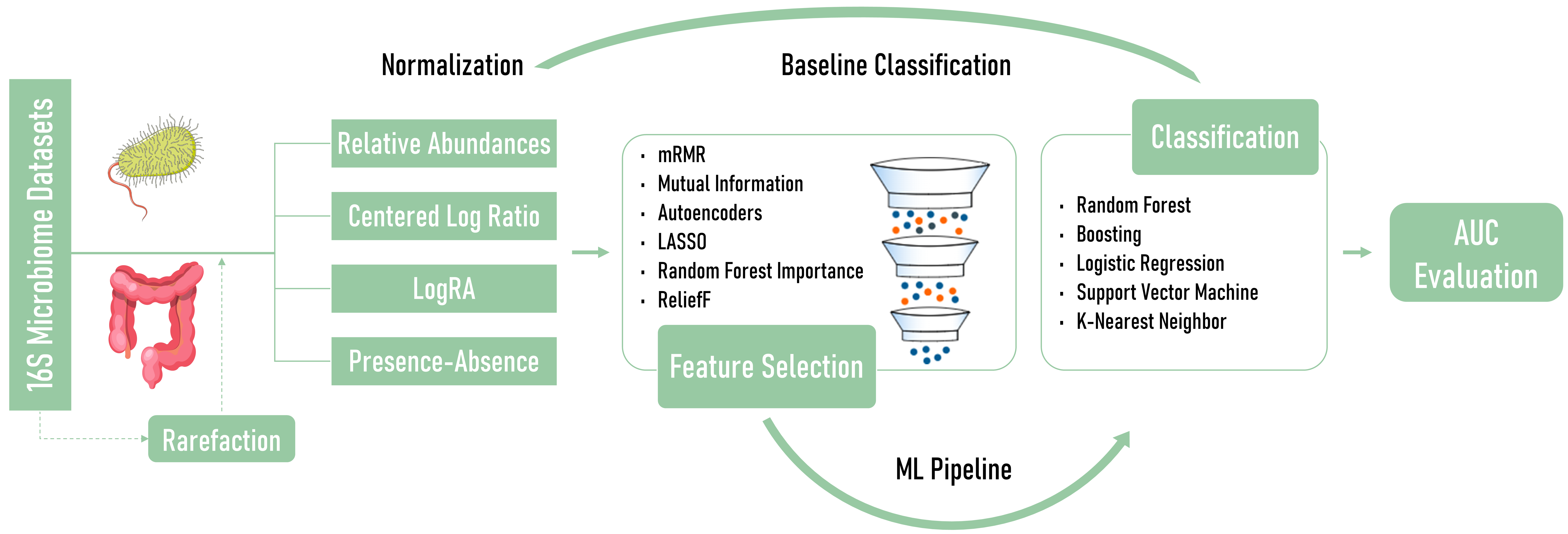
Code and supporting data for the article: "Exploring the role of normalization and feature selection in microbiome disease classification pipelines."
The repository contains the following folders:
- 1. data: contains OTU/ASV tables and class annotations for the 15 curated datasets considered.
- 2. src: code writen to perform the analyses from the article and the statistical tests
- 3. results: tables containing global nested cross validation results
- 4. figures
License: This project is licensed under GNU GPL 3.0 - check LICENSE file for more details.
| Dataset | Samples (Cases, Controls) | Features | IR | Project ID |
|---|---|---|---|---|
| ART | 114 (86, 28) | 10733 | 3.07 | PRJNA203810 |
| CDI | 336 (93, 243) | 3456 | 2.61 | 10.1128/mbio.01021-14 (DOI) |
| CRC1 | 490 (229, 261) | 6920 | 1.14 | PRJNA290926 |
| CRC2 | 102 (46, 56) | 837 | 1.22 | SRP005150 |
| HIV | 350 (293, 57) | 14425 | 5.14 | PRJNA307231 |
| CD1 | 140 (78, 62) | 3547 | 1.26 | PRJNA237362 |
| CD2 | 160 (68, 92) | 3547 | 1.35 | PRJNA237362 |
| IBD1 | 91 (67, 24) | 2742 | 2.79 | PRJNA82109 |
| IBD2 | 114 (68, 46) | 1496 | 1.48 | 10.1053/j.gastro.2010.08.049 (DOI) |
| CIR | 77 (51, 26) | 3104 | 1.96 | PRJNA174838 |
| MHE | 77 (26, 51) | 3104 | 1.96 | PRJNA174838 |
| OB | 281 (220, 61) | 6386 | 3.61 | PRJNA32089 |
| PAR1 | 148 (74, 74) | 10232 | 1.00 | PRJEB4927 |
| PAR2 | 333 (201, 132) | 6844 | 1.52 | PRJNA601994 |
| PAR3 | 507 (323, 184) | 12198 | 1.76 | PRJNA601994 |
Notes:
- ART: Arthritis; CDI: Clostridium difficile Infection; CRC1 and CRC2: Colorectal Cancer; HIV: Human Immunodeficiency Virus; CD1 and CD2: Crohn's Disease; IBD1 and IBD2: Inflammatory Bowel Disease; CIR: Cirrhosis; MHE: Minimal Hepatic Encephalopathy; OB: Obesity; PAR1, PAR2, and PAR3: Parkinson's Disease.
- CD1 and CD2 were taken from MLRepo, PAR2 and PAR3 were retrieved from their respective article sources, and the remaining datasets were obtained from MicrobiomeHD.
- Project IDs from NCBI for raw data. IBD2 data is only available via MicrobiomeHD repository, CDI raw data is available at mothur (https://mothur.org/CDI_MicrobiomeModeling/)
Version History
main @ b9472f1 (earliest) Created 13th Jul 2025 at 17:40 by Ignacio Garach Vélez
Add files via upload
Frozen
 main
mainb9472f1
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Garach, I. (2025). Exploring the role of normalization and feature selection in microbiome disease classification pipelines. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.1807.1
Activity
Views: 492 Downloads: 113
Created: 13th Jul 2025 at 17:40
Annotated Properties
Topic annotations
 Attributions
AttributionsNone
 View on GitHub
View on GitHub https://orcid.org/0009-0006-6618-7949
https://orcid.org/0009-0006-6618-7949