Workflow Type: Galaxy
Open
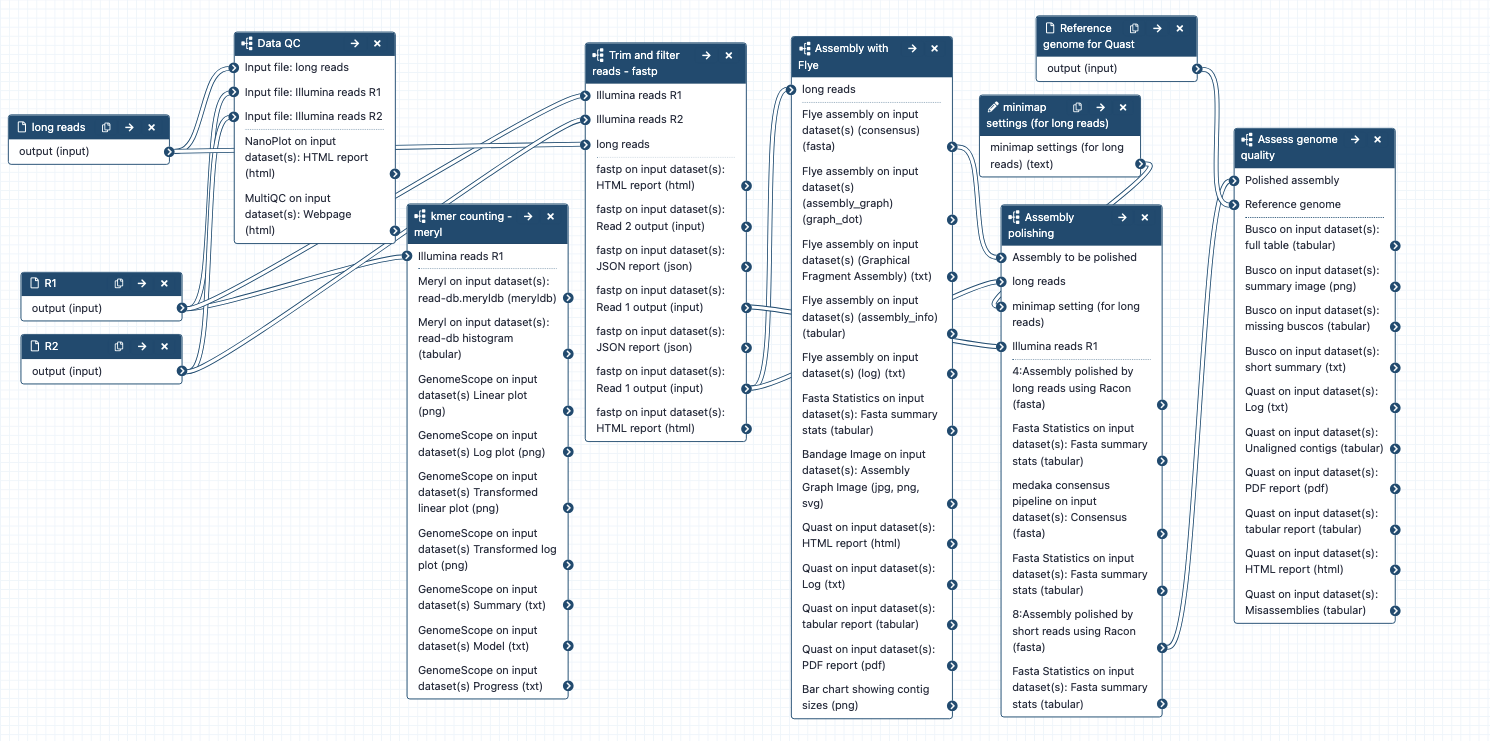
Combined workflow for large genome assembly
The tutorial document for this workflow is here: https://doi.org/10.5281/zenodo.5655813
What it does: A workflow for genome assembly, containing subworkflows:
- Data QC
- Kmer counting
- Trim and filter reads
- Assembly with Flye
- Assembly polishing
- Assess genome quality
Inputs:
- long reads and short reads in fastq format
- reference genome for Quast
Outputs:
- Data information - QC, kmers
- Filtered, trimmed reads
- Genome assembly, assembly graph, stats
- Polished assembly, stats
- Quality metrics - Busco, Quast
Options
- Omit some steps - e.g. Data QC and kmer counting
- Replace a module with one using a different tool - e.g. change assembly tool
Infrastructure_deployment_metadata: Galaxy Australia (Galaxy)
Steps
| ID | Name | Description |
|---|---|---|
| 5 | kmer counting - meryl | c0b5fbb61b12154d |
| 6 | Data QC | ae3b48869cad659b |
| 7 | Trim and filter reads - fastp | 2ed2445df255451a |
| 8 | Assembly with Flye | ec0fc1dbb6a13fe5 |
| 9 | Assembly polishing | 1fbcdce7d5823b15 |
| 10 | Assess genome quality | 9d0e6aba18484ea8 |
Version History
Version 1 (earliest) Created 8th Nov 2021 at 06:08 by Anna Syme
Added/updated 2 files
Open
 master
master5ce10d2
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Citation
Syme, A. (2021). Combined workflows for large genome assembly. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.230.1
License
Activity
Views: 7566 Downloads: 758 Runs: 14
Created: 8th Nov 2021 at 06:08
Last updated: 11th Nov 2021 at 03:37
 Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673