Workflow Type: Common Workflow Language
Open
Stable
Author: AMBARISH KUMAR er.ambarish@gmail.com; ambari73_sit@jnu.ac.in
This is a proposed standard operating procedure for genomic variant detection using VARSCAN.
It is hoped to be effective and useful for getting SARS-CoV-2 genome variants.
It uses Illumina RNASEQ reads and genome sequence.
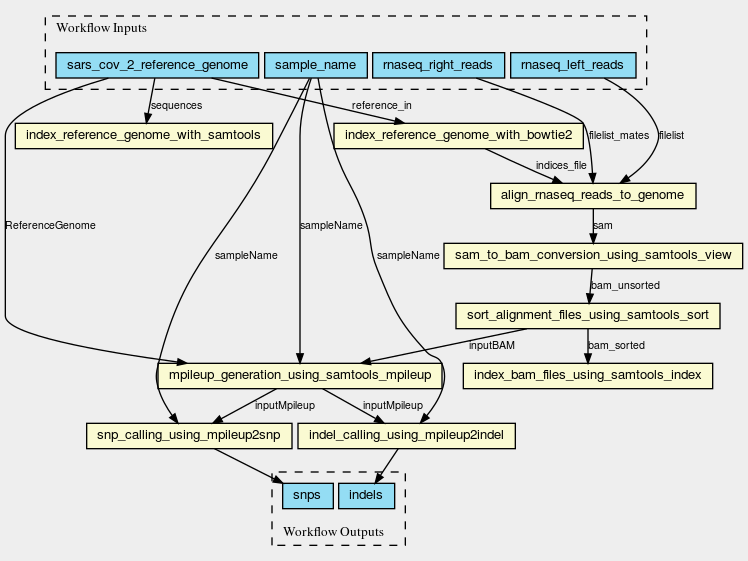
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| sars_cov_2_reference_genome | n/a | n/a |
|
| rnaseq_left_reads | n/a | n/a |
|
| rnaseq_right_reads | n/a | n/a |
|
| sample_name | n/a | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| index_reference_genome_with_bowtie2 | n/a | n/a |
| align_rnaseq_reads_to_genome | n/a | n/a |
| index_reference_genome_with_samtools | n/a | n/a |
| sam_to_bam_conversion_using_samtools_view | n/a | n/a |
| sort_alignment_files_using_samtools_sort | n/a | n/a |
| index_bam_files_using_samtools_index | n/a | n/a |
| mpileup_generation_using_samtools_mpileup | n/a | n/a |
| snp_calling_using_mpileup2snp | n/a | n/a |
| indel_calling_using_mpileup2indel | n/a | n/a |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| snps | n/a | n/a |
|
| indels | n/a | n/a |
|
Version History
Version 1 (earliest) Created 17th Jun 2020 at 07:24 by Ambarish Kumar
Added/updated 2 files
Open
 master
masterdc0b2e7
 Creators and Submitter
Creators and SubmitterCreator
Submitter
License
Activity
Views: 4500 Downloads: 1037
Created: 17th Jun 2020 at 07:24
 Attributions
AttributionsNone
 View on GitHub
View on GitHub External Link
External Link