Mutation Free Energy Calculations using BioExcel Building Blocks (biobb)
Based on the official pmx tutorial.
This tutorial aims to illustrate how to compute a fast-growth mutation free energy calculation, step by step, using the BioExcel Building Blocks library (biobb). The particular example used is the Staphylococcal nuclease protein (PDB code 1STN), a small, minimal protein, appropriate for a short tutorial.
The non-equilibrium free energy calculation protocol performs a fast alchemical transition in the direction WT->Mut and back Mut->WT. The two equilibrium trajectories needed for the tutorial, one for Wild Type (WT) and another for the Mutated (Mut) protein (Isoleucine 10 to Alanine -I10A-), have already been generated and are included in this example. We will name WT as stateA and Mut as stateB.
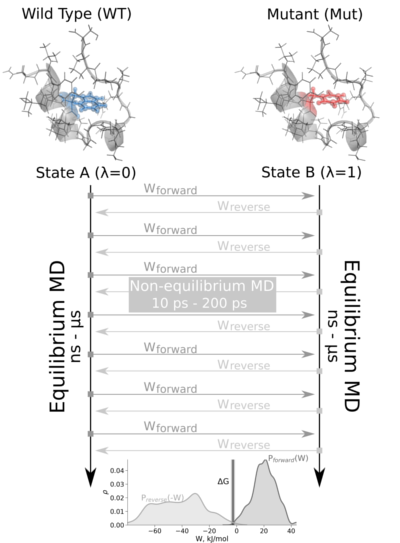
The tutorial calculates the free energy difference in the folded state of a protein. Starting from two 1ns-length independent equilibrium simulations (WT and mutant), snapshots are selected to start fast (50ps) transitions driving the system in the forward (WT to mutant) and reverse (mutant to WT) directions, and the work values required to perform these transitions are collected. With these values, Crooks Gaussian Intersection (CGI), Bennett Acceptance Ratio (BAR) and Jarzynski estimator methods are used to calculate the free energy difference between the two states.
Please note that for the sake of disk space this tutorial is using 1ns-length equilibrium trajectories, whereas in the original example the equilibrium trajectories used were obtained from 10ns-length simulations.
Copyright & Licensing
This software has been developed in the MMB group at the BSC & IRB for the European BioExcel, funded by the European Commission (EU H2020 823830, EU H2020 675728).
- (c) 2015-2022 Barcelona Supercomputing Center
- (c) 2015-2022 Institute for Research in Biomedicine
Licensed under the Apache License 2.0, see the file LICENSE for details.

Version History
Version 7 (latest) Created 6th Aug 2025 at 09:42 by Genís Bayarri
Update to BioBB 4.2.*
Frozen
 Version-7
Version-7bc79de5
Version 6 Created 4th Mar 2024 at 14:26 by Genís Bayarri
Update to BioBB 4.1.*
Frozen
 Version-6
Version-63706360
Version 5 Created 26th Jul 2023 at 10:23 by Genís Bayarri
Fixed importlib_metadata bug
Frozen
 Version-5
Version-5098c09d
Version 4 Created 16th Sep 2022 at 10:35 by Genís Bayarri
Update to BioBB 3.8.*. taken from Git commit 5b20069
Frozen
 Version-4
Version-4a9f9ac3
Version 3 Created 1st Jul 2021 at 11:33 by Douglas Lowe
Updated to BioBB 3.6.0. Taken from Git commit f329e83
Frozen
 Version-3
Version-38659eeb
Version 2 Created 7th May 2021 at 14:22 by Douglas Lowe
Updated for GROMACS 2020.4 and BioExcel Building Blocks version 3.5. Taken from Git commit ba3c37d
Frozen
 master
masterf14839e
Version 1 (earliest) Created 15th Sep 2020 at 12:56 by Douglas Lowe
Initial commit. Taken from Git commit 3272c52
Frozen
 master
masterd778945
 Creators and Submitter
Creators and SubmitterCreators
Submitter
Views: 10850 Downloads: 3435
Created: 15th Sep 2020 at 12:56
Last updated: 6th Aug 2025 at 09:42
 Tags
TagsThis item has not yet been tagged.
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub https://orcid.org/0000-0003-0513-0288
https://orcid.org/0000-0003-0513-0288