Workflow Type: Galaxy
Frozen
Stable
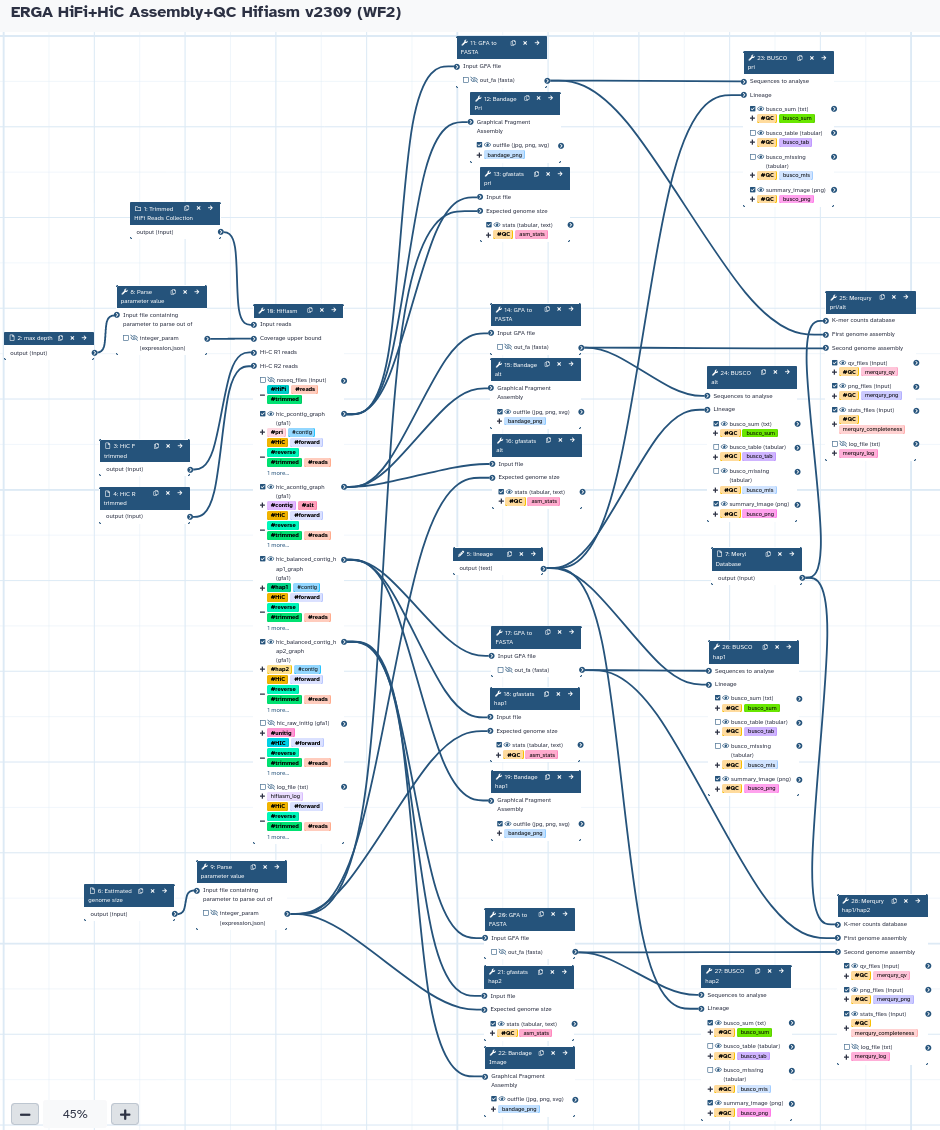
The workflow takes a trimmed HiFi reads collection, Forward/Reverse HiC reads, and the max coverage depth (calculated from WF1) to run Hifiasm in HiC phasing mode. It produces both Pri/Alt and Hap1/Hap2 assemblies, and runs all the QC analysis (gfastats, BUSCO, and Merqury). The default Hifiasm purge level is Light (l1).
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Estimated genome size | #main/Estimated genome size | n/a |
|
| HiC F trimmed | #main/HiC F trimmed | n/a |
|
| HiC R trimmed | #main/HiC R trimmed | n/a |
|
| Meryl Database | #main/Meryl Database | n/a |
|
| Trimmed HiFi Reads Collection | #main/Trimmed HiFi Reads Collection | Collection of Long reads in fastq format. |
|
| lineage | #main/lineage | lineage for BUSCO, e.g.: arthropoda_odb10, vertebrata_odb10, mammalia_odb10, aves_odb10, tetrapoda_odb10 ... |
|
| max depth | #main/max depth | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 7 | Parse parameter value | param_value_from_file |
| 8 | Parse parameter value | param_value_from_file |
| 9 | Hifiasm | toolshed.g2.bx.psu.edu/repos/bgruening/hifiasm/hifiasm/0.19.5+galaxy0 |
| 10 | GFA to FASTA | toolshed.g2.bx.psu.edu/repos/iuc/gfa_to_fa/gfa_to_fa/0.1.2 |
| 11 | Bandage Pri | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 12 | gfastats pri | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.5+galaxy0 |
| 13 | GFA to FASTA | toolshed.g2.bx.psu.edu/repos/iuc/gfa_to_fa/gfa_to_fa/0.1.2 |
| 14 | Bandage alt | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 15 | gfastats alt | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.6+galaxy0 |
| 16 | GFA to FASTA | toolshed.g2.bx.psu.edu/repos/iuc/gfa_to_fa/gfa_to_fa/0.1.2 |
| 17 | gfastats hap1 | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.6+galaxy0 |
| 18 | Bandage hap1 | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 19 | GFA to FASTA | toolshed.g2.bx.psu.edu/repos/iuc/gfa_to_fa/gfa_to_fa/0.1.2 |
| 20 | gfastats hap2 | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.5+galaxy0 |
| 21 | Bandage Image | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 22 | BUSCO pri | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.3.2+galaxy0 |
| 23 | BUSCO alt | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.3.2+galaxy0 |
| 24 | Merqury pri/alt | toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy2 |
| 25 | BUSCO hap1 | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.3.2+galaxy0 |
| 26 | BUSCO hap2 | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.3.2+galaxy0 |
| 27 | Merqury hap1/hap2 | toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_22 | #main/_anonymous_output_22 | n/a |
|
| _anonymous_output_23 | #main/_anonymous_output_23 | n/a |
|
| _anonymous_output_24 | #main/_anonymous_output_24 | n/a |
|
| _anonymous_output_25 | #main/_anonymous_output_25 | n/a |
|
| _anonymous_output_26 | #main/_anonymous_output_26 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
Version 1 (earliest) Created 9th Oct 2023 at 13:47 by Diego De Panis
Initial commit
Frozen
 Version-1
Version-1958e932
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
ERGA
Submitter
Discussion Channel
Citation
De Panis, D. (2024). ERGA HiFi+HiC Assembly+QC Hifiasm v2309 (WF2). WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.605.1
License
Activity
Views: 6770 Downloads: 575 Runs: 7
Created: 9th Oct 2023 at 13:47
Annotated Properties
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-3679-9585
https://orcid.org/0000-0002-3679-9585