Workflow Type: Galaxy
Frozen
This is part of a series of workflows to annotate a genome, tagged with TSI-annotation.
These workflows are based on command-line code by Luke Silver, converted into Galaxy Australia workflows.
The workflows can be run in this order:
- Repeat masking
- RNAseq QC and read trimming
- Find transcripts
- Combine transcripts
- Extract transcripts
- Convert formats
- Fgenesh annotation
About this workflow:
- Run this workflow per tissue.
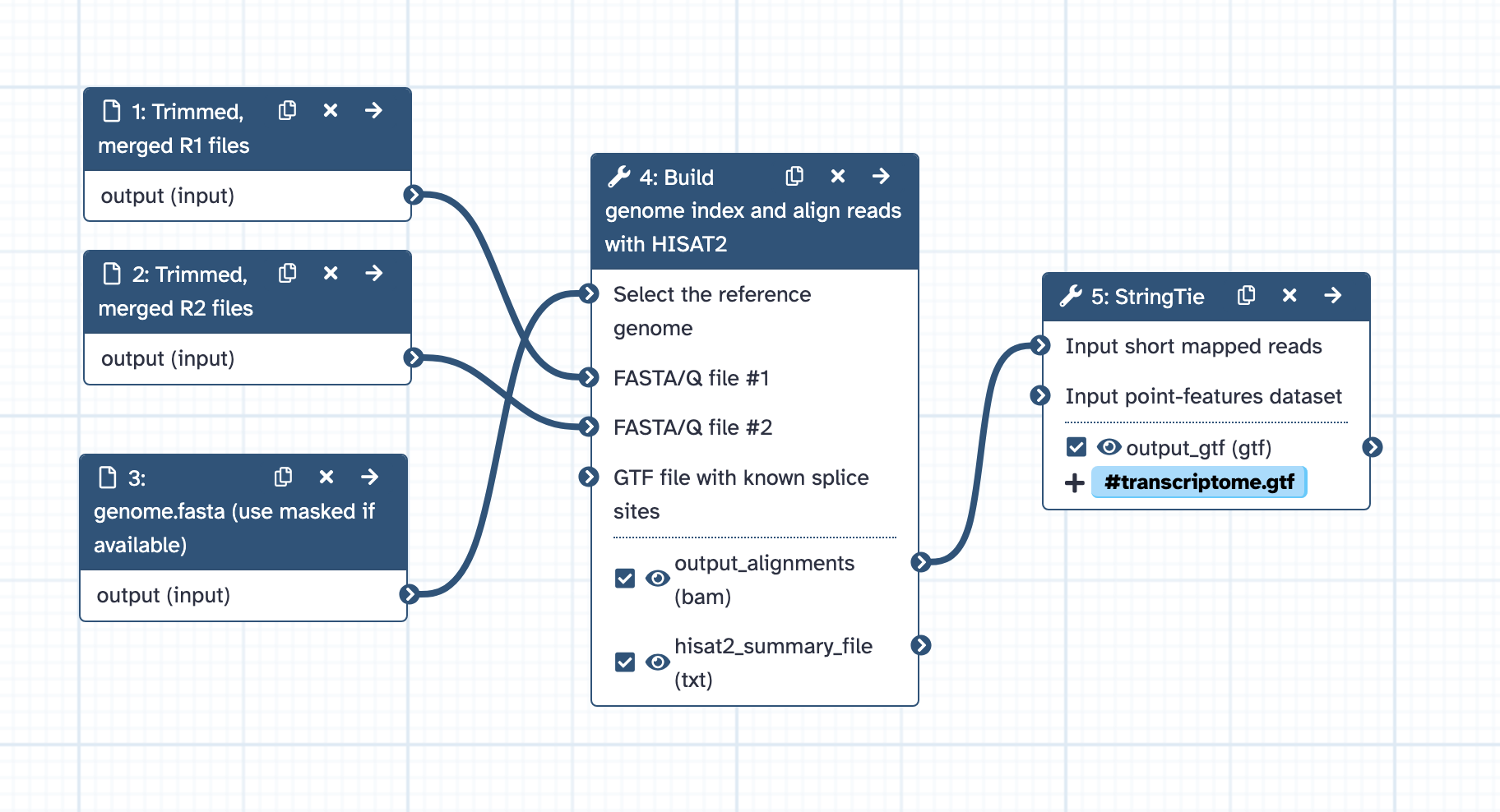
- Inputs: masked_genome.fasta and the trimmed RNAseq reads (R1 and R2) from one type of tissue.
- Index genome and align reads to genome with HISAT2, with default settings except for: Advanced options: spliced alignment options: specify options: Transcriptome assembly reporting: selected option: Report alignments tailored for transcript assemblers including StringTie (equivalent to -dta flag).
- Runs samtools sort to sort bam by coordinate.
- Runs StringTie to generate gtf from sorted bam.
- Output: transcripts.gtf from a single tissue.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Trimmed, merged R1 files | #main/Trimmed, merged R1 files | n/a |
|
| Trimmed, merged R2 files | #main/Trimmed, merged R2 files | n/a |
|
| genome.fasta (use masked if available) | #main/genome.fasta (use masked if available) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Build genome index and align reads with HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.2.1+galaxy1 |
| 4 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/2.2.1+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| hisat2_summary_file | #main/hisat2_summary_file | n/a |
|
| output_alignments | #main/output_alignments | n/a |
|
| output_gtf | #main/output_gtf | n/a |
|
Version History
Version 1 (earliest) Created 8th May 2024 at 07:51 by Anna Syme
Initial commit
Frozen
 Version-1
Version-149078a8
 Creators and Submitter
Creators and SubmitterCreators
Submitter
Citation
Silver, L., & Syme, A. (2024). Find transcripts - TSI. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.877.1
Activity
Views: 4590 Downloads: 518 Runs: 1
Created: 8th May 2024 at 07:51
Last updated: 9th May 2024 at 05:04
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673