Workflow Type: Galaxy
Frozen
This is part of a series of workflows to annotate a genome, tagged with TSI-annotation.
These workflows are based on command-line code by Luke Silver, converted into Galaxy Australia workflows.
The workflows can be run in this order:
- Repeat masking
- RNAseq QC and read trimming
- Find transcripts
- Combine transcripts
- Extract transcripts
- Convert formats
- Fgenesh annotation
About this workflow:
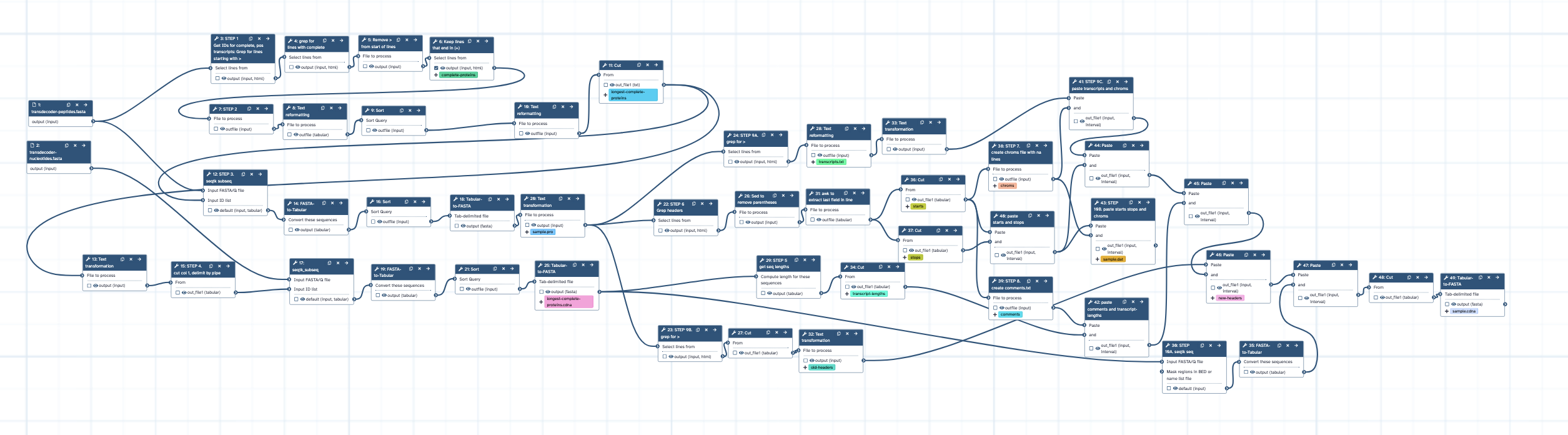
- Inputs: transdecoder-peptides.fasta, transdecoder-nucleotides.fasta
- Runs many steps to convert outputs into the formats required for Fgenesh - .pro, .dat and .cdna
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| transdecoder-nucleotides.fasta | transdecoder-nucleotides.fasta | n/a |
|
| transdecoder-peptides.fasta | transdecoder-peptides.fasta | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | STEP 1 Get IDs for complete, pos transcripts: Grep for lines starting with > | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy0 |
| 3 | grep for lines with complete | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy0 |
| 4 | Remove > from start of lines | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 5 | Keep lines that end in (+) | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy0 |
| 6 | STEP 2 | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy0 |
| 7 | Text reformatting | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy0 |
| 8 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.3+galaxy0 |
| 9 | Text reformatting | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy0 |
| 10 | Cut | Cut1 |
| 11 | STEP 3. seqtk subseq | toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_subseq/1.4+galaxy0 |
| 12 | Text transformation | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 13 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 14 | STEP 4. cut col 1, delimit by pipe | Cut1 |
| 15 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.3+galaxy0 |
| 16 | seqtk_subseq | toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_subseq/1.4+galaxy0 |
| 17 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 18 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 19 | Text transformation | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 20 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.3+galaxy0 |
| 21 | STEP 6 Grep headers | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy0 |
| 22 | STEP 9B. grep for > | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy0 |
| 23 | STEP 9A. grep for > | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy0 |
| 24 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 25 | Sed to remove parentheses | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 26 | Cut | Cut1 |
| 27 | Text reformatting | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy0 |
| 28 | STEP 5 get seq lengths | toolshed.g2.bx.psu.edu/repos/devteam/fasta_compute_length/fasta_compute_length/1.0.3 |
| 29 | STEP 10A. seqtk seq | toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_seq/1.4+galaxy0 |
| 30 | awk to extract last field in line | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy0 |
| 31 | Text transformation | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 32 | Text transformation | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/9.3+galaxy0 |
| 33 | Cut | Cut1 |
| 34 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 35 | Cut | Cut1 |
| 36 | Cut | Cut1 |
| 37 | STEP 7. create chroms file with na lines | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_line/9.3+galaxy0 |
| 38 | STEP 8. create comments.txt | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_line/9.3+galaxy0 |
| 39 | paste starts and stops | Paste1 |
| 40 | STEP 9C. paste transcripts and chroms | Paste1 |
| 41 | paste comments and transcript-lengths | Paste1 |
| 42 | STEP 10B. paste starts stops and chroms | Paste1 |
| 43 | Paste | Paste1 |
| 44 | Paste | Paste1 |
| 45 | Paste | Paste1 |
| 46 | Paste | Paste1 |
| 47 | Cut | Cut1 |
| 48 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| output | output | n/a |
|
Version History
Version 1 (earliest) Created 8th May 2024 at 08:23 by Anna Syme
Initial commit
Frozen
 Version-1
Version-1caf683f
 Creators and Submitter
Creators and SubmitterCreators
Submitter
Tools
Citation
Silver, L., & Syme, A. (2024). Convert formats - TSI. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.880.1
Activity
Views: 4693 Downloads: 650 Runs: 1
Created: 8th May 2024 at 08:23
Last updated: 9th May 2024 at 05:09
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-9906-0673
https://orcid.org/0000-0002-9906-0673