Workflows
What is a Workflow?Filters
WorkflowHub Knowledge Graph
A tool to generate a knowledge graph from a source of RO Crates. By default, this tool sources and generates an RDF graph of crates from WorkflowHub.
Type: Snakemake
Creators: Alexander Hambley, Oliver Woolland, Eli Chadwick, Volodymyr Savchenko, José Mª Fernández, Stian Soiland-Reyes
Submitter: Eli Chadwick
COnSensus Interaction Network InFErence Service
Inference framework for reconstructing networks using a consensus approach between multiple methods and data sources.

Reference
[Manica, Matteo, Charlotte, Bunne, Roland, Mathis, Joris, Cadow, Mehmet Eren, Ahsen, Gustavo A, Stolovitzky, and María Rodríguez, Martínez. "COSIFER: a python package for the consensus inference of molecular interaction ...
Type: Common Workflow Language
Creators: Laura Rodriguez-Navas, José Mª Fernández
Submitter: Laura Rodriguez-Navas
Joint multi-omics dimensionality reduction approaches for CAKUT data using peptidome and proteome data
Brief description In (Cantini et al. 2020), Cantini et al. evaluated 9 representative joint dimensionality reduction (jDR) methods for multi-omics integration and analysis and . The methods are Regularized Generalized Canonical Correlation Analysis (RGCCA), Multiple co-inertia analysis (MCIA), Multi-Omics Factor Analysis (MOFA), Multi-Study Factor Analysis (MSFA), iCluster, Integrative NMF ...
Type: Snakemake
Creators: Ozan Ozisik, Juma Bayjan, Cenna Doornbos, Friederike Ehrhart, Matthias Haimel, Laura Rodriguez-Navas, José Mª Fernández, Eleni Mina, Daniël Wijnbergen
Submitter: Juma Bayjan
Description
The workflow takes an input file with Cancer Driver Genes predictions (i.e. the results provided by a participant), computes a set of metrics, and compares them against the data currently stored in OpenEBench within the TCGA community. Two assessment metrics are provided for that predictions. Also, some plots (which are optional) that allow to visualize the performance of the tool are generated. The workflow consists in three standard steps, defined by OpenEBench. The tools needed ...
Type: Nextflow
Creators: José Mª Fernández, Asier Gonzalez-Uriarte, Javier Garrayo-Ventas
Submitter: Laura Rodriguez-Navas
Rare disease researchers workflow is that they submit their raw data (fastq), run the mapping and variant calling RD-Connect pipeline and obtain unannotated gvcf files to further submit to the RD-Connect GPAP or analyse on their own.
This demonstrator focuses on the variant calling pipeline. The raw genomic data is processed using the RD-Connect pipeline (Laurie et al., 2016) running on the standards (GA4GH) compliant, interoperable container ...
COnSensus Interaction Network InFErence Service
Inference framework for reconstructing networks using a consensus approach between multiple methods and data sources.
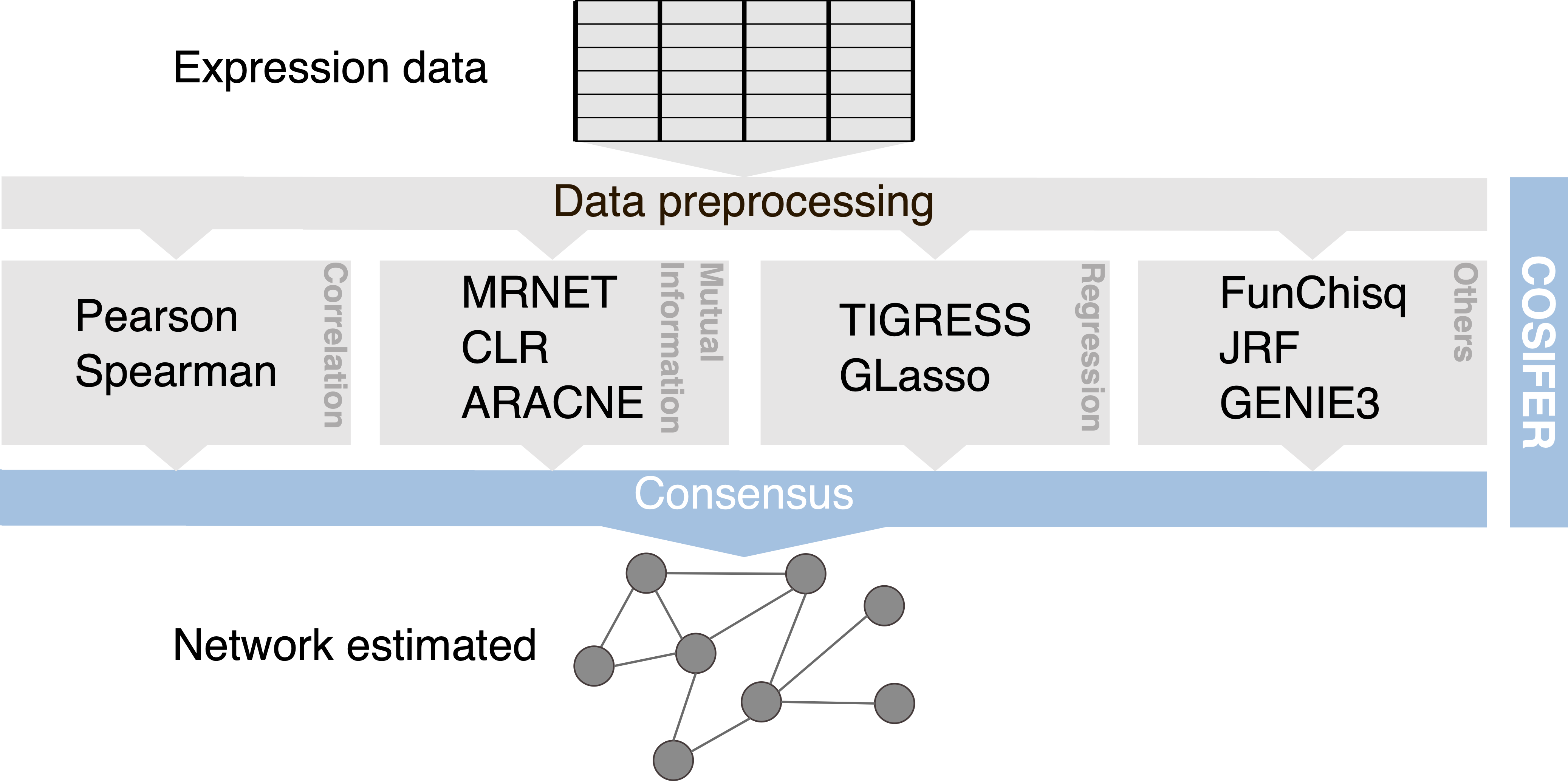
Reference
[Manica, Matteo, Charlotte, Bunne, Roland, Mathis, Joris, Cadow, Mehmet Eren, Ahsen, Gustavo A, Stolovitzky, and María Rodríguez, Martínez. "COSIFER: a python package for the consensus inference of molecular ...
Rare disease researchers workflow is that they submit their raw data (fastq), run the mapping and variant calling RD-Connect pipeline and obtain unannotated gvcf files to further submit to the RD-Connect GPAP or analyse on their own.
This demonstrator focuses on the variant calling pipeline. The raw genomic data is processed using the RD-Connect pipeline (Laurie et al., 2016) running on the standards (GA4GH) compliant, interoperable container ...
Type: Common Workflow Language
Creators: José Mª Fernández, Laura Rodriguez-Navas
Submitter: Laura Rodriguez-Navas
 Download
Download