Workflow Type: Galaxy
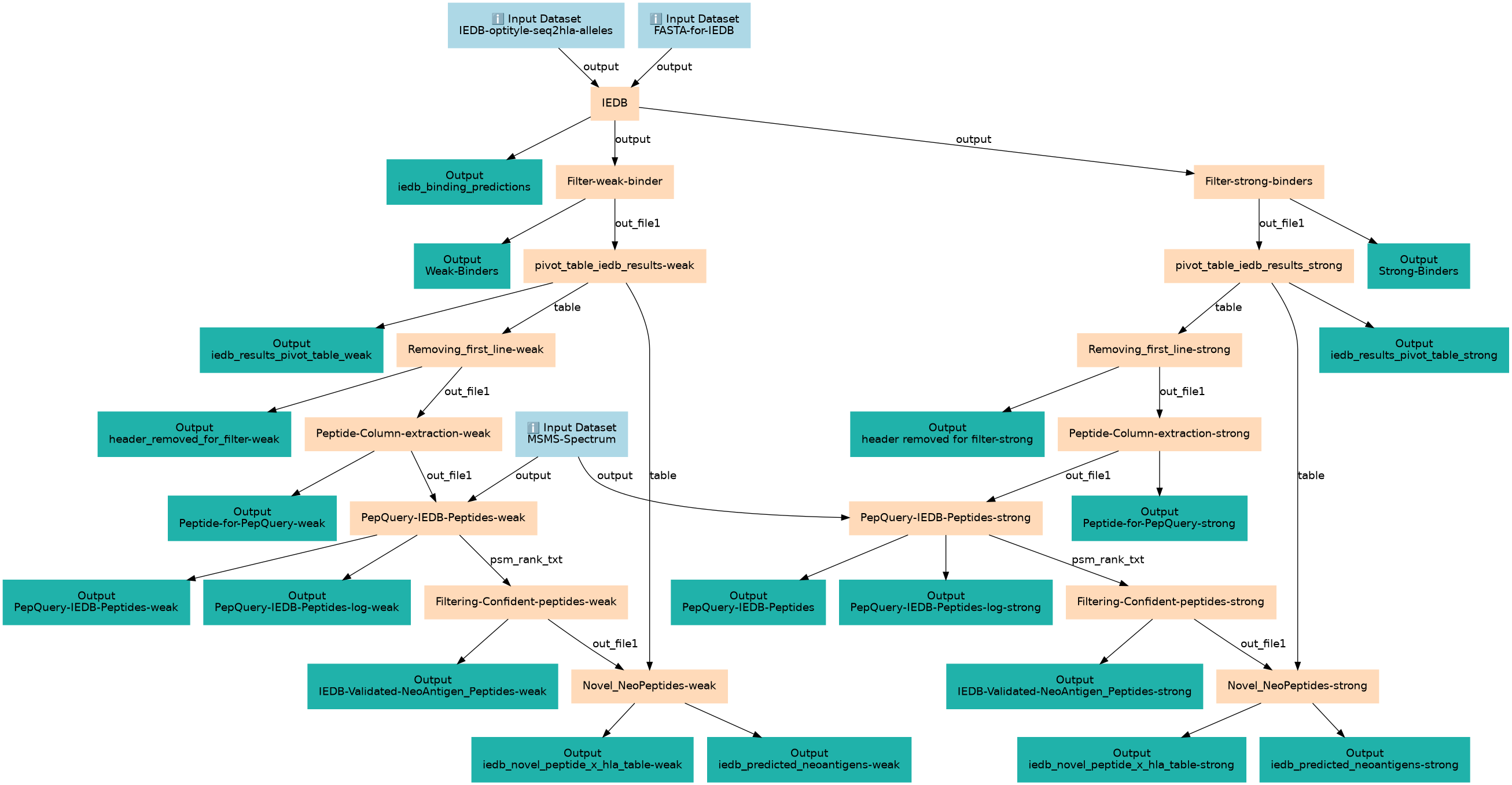
Predict binding using IEDB and check novelty peptides with PepQuery
Associated Tutorial
This workflows is part of the tutorial Neoantigen 7: IEDB binding PepQuery Validated Neopeptides, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): GalaxyP
Tutorial Author(s): Subina Mehta, Katherine Do, James Johnson
Tutorial Contributor(s): Pratik Jagtap, Timothy J. Griffin, Subina Mehta
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| FASTA-for-IEDB | #main/FASTA-for-IEDB | A fasta file of peptides to test for HLA binding, can be from Arriba |
|
| IEDB-optityle-seq2hla-alleles | #main/IEDB-optityle-seq2hla-alleles | A single column tabular file with an HLA per line IEDB format: e.g. HLA-A*01:01 |
|
| MSMS-Spectrum | #main/MSMS-Spectrum | MSMS spectrum |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | IEDB | IEDB binding prediction toolshed.g2.bx.psu.edu/repos/iuc/iedb_api/iedb_api/2.15.2 |
| 4 | Filter-weak-binder | anything less than 2 and more than 0.5 percentile rank Filter1 |
| 5 | Filter-strong-binders | anything less than 0.5 percentile rank Filter1 |
| 6 | pivot_table_iedb_results-weak | extracting allele results using pivot toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy0 |
| 7 | pivot_table_iedb_results_strong | extracting allele results using pivot toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy0 |
| 8 | Removing_first_line-weak | this is to remove header Remove beginning1 |
| 9 | Removing_first_line-strong | removing header Remove beginning1 |
| 10 | Peptide-Column-extraction-weak | cutting first column to extract peptides Cut1 |
| 11 | Peptide-Column-extraction-strong | cutting first column to extract peptides Cut1 |
| 12 | PepQuery-IEDB-Peptides-weak | Test if the peptides are novel, and search for peptide in Mass Spec results. toolshed.g2.bx.psu.edu/repos/galaxyp/pepquery2/pepquery2/2.0.2+galaxy2 |
| 13 | PepQuery-IEDB-Peptides-strong | Test if the peptides are novel, and search for peptide in Mass Spec results. toolshed.g2.bx.psu.edu/repos/galaxyp/pepquery2/pepquery2/2.0.2+galaxy2 |
| 14 | Filtering-Confident-peptides-weak | IEDB-Validated-NeoAntigen_Peptides, filtering if the validation column is yes or no Filter1 |
| 15 | Filtering-Confident-peptides-strong | IEDB-Validated-NeoAntigen_Peptides, filtering if the validation column is yes or no Filter1 |
| 16 | Novel_NeoPeptides-weak | Remove reference peptides from IeDB Pivot Table toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 17 | Novel_NeoPeptides-strong | Remove reference peptides from IeDB Pivot Table toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| IEDB-Validated-NeoAntigen_Peptides-strong | #main/IEDB-Validated-NeoAntigen_Peptides-strong | n/a |
|
| IEDB-Validated-NeoAntigen_Peptides-weak | #main/IEDB-Validated-NeoAntigen_Peptides-weak | n/a |
|
| PepQuery-IEDB-Peptides | #main/PepQuery-IEDB-Peptides | n/a |
|
| PepQuery-IEDB-Peptides-log-strong | #main/PepQuery-IEDB-Peptides-log-strong | n/a |
|
| PepQuery-IEDB-Peptides-log-weak | #main/PepQuery-IEDB-Peptides-log-weak | n/a |
|
| PepQuery-IEDB-Peptides-weak | #main/PepQuery-IEDB-Peptides-weak | n/a |
|
| Peptide-for-PepQuery-strong | #main/Peptide-for-PepQuery-strong | n/a |
|
| Peptide-for-PepQuery-weak | #main/Peptide-for-PepQuery-weak | n/a |
|
| Strong-Binders | #main/Strong-Binders | n/a |
|
| Weak-Binders | #main/Weak-Binders | n/a |
|
| header removed for filter-strong | #main/header removed for filter-strong | n/a |
|
| header_removed_for_filter-weak | #main/header_removed_for_filter-weak | n/a |
|
| iedb_binding_predictions | #main/iedb_binding_predictions | n/a |
|
| iedb_novel_peptide_x_hla_table-strong | #main/iedb_novel_peptide_x_hla_table-strong | n/a |
|
| iedb_novel_peptide_x_hla_table-weak | #main/iedb_novel_peptide_x_hla_table-weak | n/a |
|
| iedb_predicted_neoantigens-strong | #main/iedb_predicted_neoantigens-strong | n/a |
|
| iedb_predicted_neoantigens-weak | #main/iedb_predicted_neoantigens-weak | n/a |
|
| iedb_results_pivot_table_strong | #main/iedb_results_pivot_table_strong | n/a |
|
| iedb_results_pivot_table_weak | #main/iedb_results_pivot_table_weak | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1240 Downloads: 198 Runs: 0
Created: 2nd Jun 2025 at 10:51
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master