Workflow Type: Galaxy
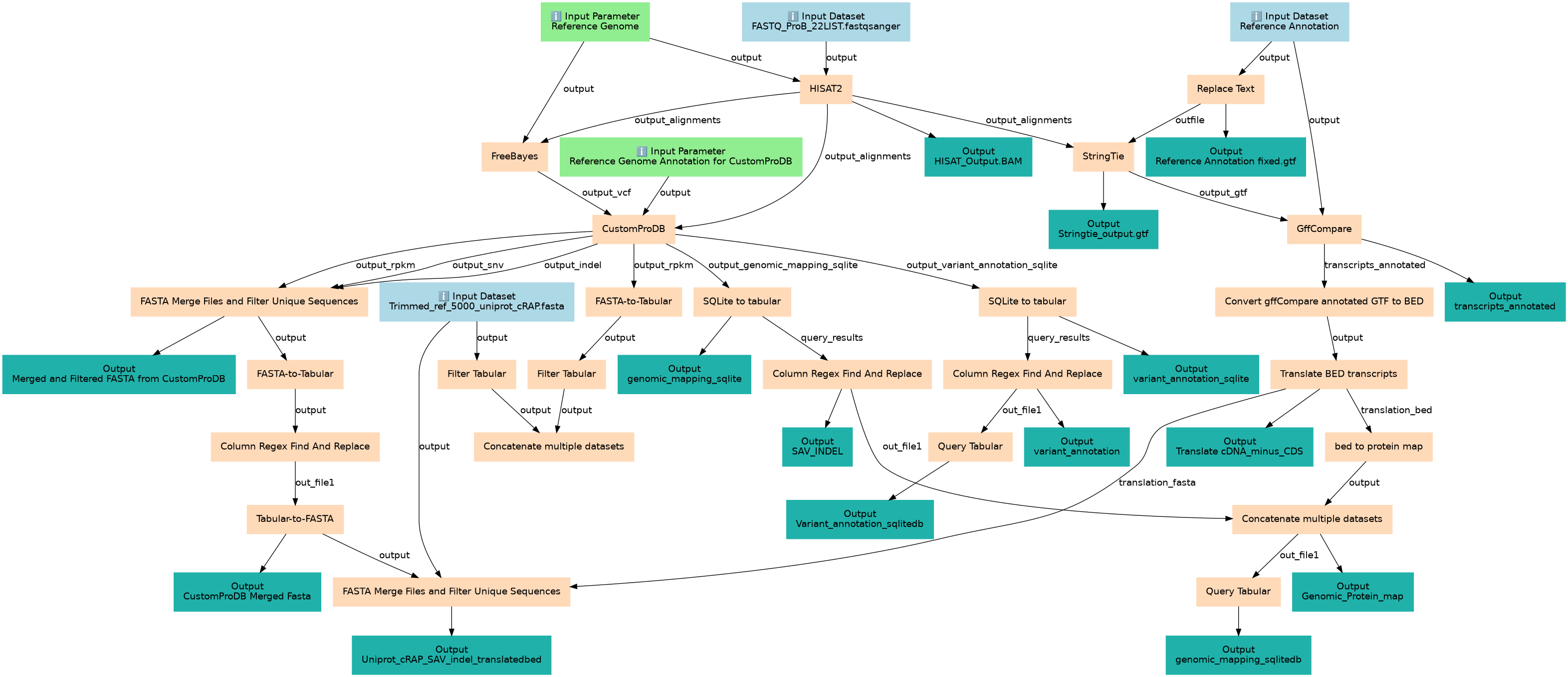
Associated Tutorial
This workflows is part of the tutorial Proteogenomics 1: Database Creation, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Tutorial Author(s): Subina Mehta, Timothy J. Griffin, Pratik Jagtap, Ray Sajulga, James Johnson, Praveen Kumar
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| FASTQ_ProB_22LIST.fastqsanger | FASTQ_ProB_22LIST.fastqsanger | n/a |
|
| Reference Annotation | Reference Annotation | n/a |
|
| Reference Genome | Reference Genome | n/a |
|
| Reference Genome Annotation for CustomProDB | Reference Genome Annotation for CustomProDB | n/a |
|
| Trimmed_ref_5000_uniprot_cRAP.fasta | Trimmed_ref_5000_uniprot_cRAP.fasta | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 5 | Filter Tabular | toolshed.g2.bx.psu.edu/repos/iuc/filter_tabular/filter_tabular/3.3.1 |
| 6 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.5+galaxy0 |
| 7 | HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.2.1+galaxy1 |
| 8 | FreeBayes | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/freebayes/1.3.9+galaxy0 |
| 9 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/2.2.3+galaxy0 |
| 10 | CustomProDB | toolshed.g2.bx.psu.edu/repos/galaxyp/custom_pro_db/custom_pro_db/1.22.0 |
| 11 | GffCompare | toolshed.g2.bx.psu.edu/repos/iuc/gffcompare/gffcompare/0.12.6+galaxy0 |
| 12 | FASTA Merge Files and Filter Unique Sequences | toolshed.g2.bx.psu.edu/repos/galaxyp/fasta_merge_files_and_filter_unique_sequences/fasta_merge_files_and_filter_unique_sequences/1.2.0 |
| 13 | SQLite to tabular | toolshed.g2.bx.psu.edu/repos/iuc/sqlite_to_tabular/sqlite_to_tabular/3.2.1 |
| 14 | SQLite to tabular | toolshed.g2.bx.psu.edu/repos/iuc/sqlite_to_tabular/sqlite_to_tabular/3.2.1 |
| 15 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 16 | Convert gffCompare annotated GTF to BED | toolshed.g2.bx.psu.edu/repos/galaxyp/gffcompare_to_bed/gffcompare_to_bed/0.2.1 |
| 17 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.1 |
| 18 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 19 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 20 | Filter Tabular | toolshed.g2.bx.psu.edu/repos/iuc/filter_tabular/filter_tabular/3.3.1 |
| 21 | Translate BED transcripts | toolshed.g2.bx.psu.edu/repos/galaxyp/translate_bed/translate_bed/0.1.0 |
| 22 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 23 | Query Tabular | toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 24 | Concatenate multiple datasets | toolshed.g2.bx.psu.edu/repos/artbio/concatenate_multiple_datasets/cat_multi_datasets/1.4.3 |
| 25 | bed to protein map | toolshed.g2.bx.psu.edu/repos/galaxyp/bed_to_protein_map/bed_to_protein_map/0.2.0 |
| 26 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 27 | Concatenate multiple datasets | toolshed.g2.bx.psu.edu/repos/artbio/concatenate_multiple_datasets/cat_multi_datasets/1.4.3 |
| 28 | FASTA Merge Files and Filter Unique Sequences | toolshed.g2.bx.psu.edu/repos/galaxyp/fasta_merge_files_and_filter_unique_sequences/fasta_merge_files_and_filter_unique_sequences/1.2.0 |
| 29 | Query Tabular | toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Reference Annotation fixed.gtf | Reference Annotation fixed.gtf | n/a |
|
| HISAT_Output.BAM | HISAT_Output.BAM | n/a |
|
| Stringtie_output.gtf | Stringtie_output.gtf | n/a |
|
| transcripts_annotated | transcripts_annotated | n/a |
|
| Merged and Filtered FASTA from CustomProDB | Merged and Filtered FASTA from CustomProDB | n/a |
|
| genomic_mapping_sqlite | genomic_mapping_sqlite | n/a |
|
| variant_annotation_sqlite | variant_annotation_sqlite | n/a |
|
| SAV_INDEL | SAV_INDEL | n/a |
|
| variant_annotation | variant_annotation | n/a |
|
| Translate cDNA_minus_CDS | Translate cDNA_minus_CDS | n/a |
|
| Variant_annotation_sqlitedb | Variant_annotation_sqlitedb | n/a |
|
| CustomProDB Merged Fasta | CustomProDB Merged Fasta | n/a |
|
| Genomic_Protein_map | Genomic_Protein_map | n/a |
|
| Uniprot_cRAP_SAV_indel_translatedbed | Uniprot_cRAP_SAV_indel_translatedbed | n/a |
|
| genomic_mapping_sqlitedb | genomic_mapping_sqlitedb | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1293 Downloads: 190 Runs: 0
Created: 2nd Jun 2025 at 10:51
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master