Workflow Type: Shell Script
Open
Work-in-progress
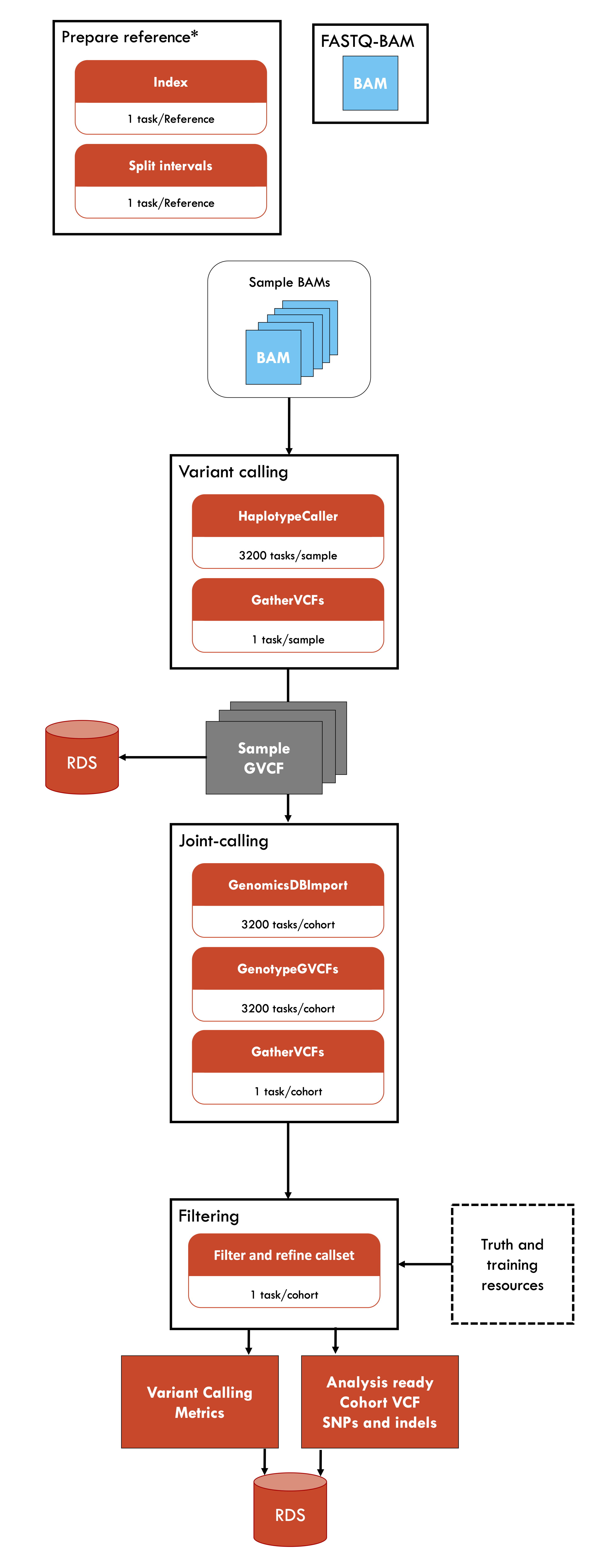
Germline-ShortV @ NCI-Gadi is an implementation of the BROAD Institute's best practice workflow for germline short variant discovery. This implementation is optimised for the National Compute Infrastucture's Gadi HPC, utilising scatter-gather parallelism to enable use of multiple nodes with high CPU or memory efficiency. This workflow requires sample BAM files, which can be generated using the Fastq-to-bam @ NCI-Gadi pipeline. Germline-ShortV can be applied to model and non-model organisms (including non-diploid organisms).
Infrastructure_deployment_metadata: Gadi (NCI)
Version History
Version 1 (earliest) Created 17th Aug 2021 at 05:35 by Tracy Chew
Added/updated 2 files
Open
 master
mastera302549
 Creators and Submitter
Creators and SubmitterCreators
Submitter
Discussion Channel
Citation
Sadsad, R., Samaha, G., & Chew, T. (2021). Germline-ShortV @ NCI-Gadi. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.143.1
Activity
Views: 5377 Downloads: 801
Created: 17th Aug 2021 at 05:35
Last updated: 25th Jul 2025 at 03:04
Annotated Properties
Topic annotations
Genomics, DNA mutation, Whole genome sequencing, Population genomics, Genetic variation, DNA polymorphism
Operation annotations
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub https://orcid.org/0000-0003-2488-953X
https://orcid.org/0000-0003-2488-953X