Workflow Type: Galaxy
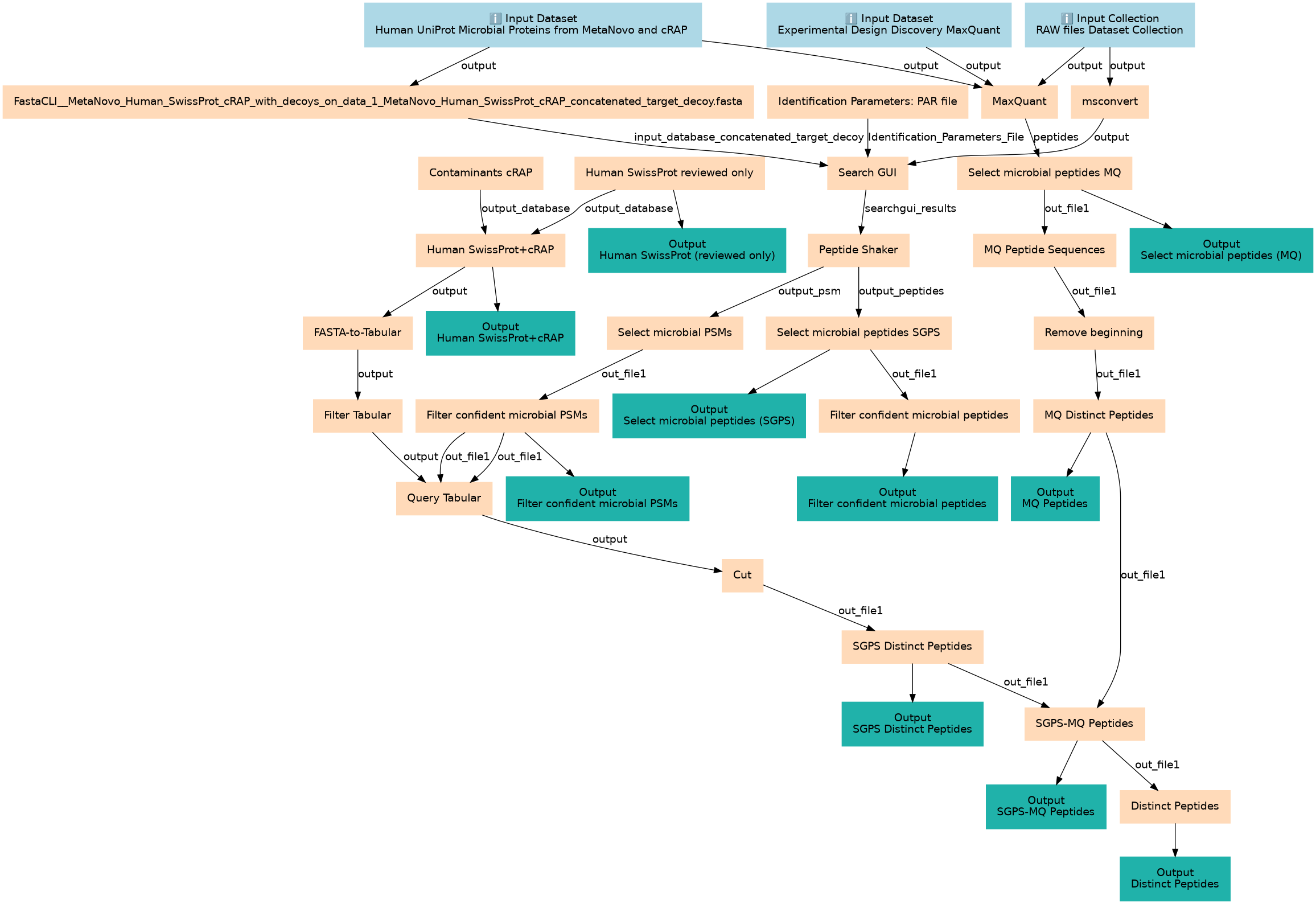
Discovery workflow with SG/PS and MaxQuant to generate microbial peptides
Associated Tutorial
This workflows is part of the tutorial Clinical Metaproteomics 2: Discovery, available in the GTN
Thanks to...
Workflow Author(s): Subina Mehta
Tutorial Author(s): Subina Mehta, Katherine Do, Dechen Bhuming
Tutorial Contributor(s): Pratik Jagtap, Timothy J. Griffin
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Experimental Design Discovery MaxQuant | Experimental Design Discovery MaxQuant | n/a |
|
| Human UniProt Microbial Proteins (from MetaNovo) and cRAP | Human UniProt Microbial Proteins (from MetaNovo) and cRAP | FastaCLI: MetaNovo Human SwissProt cRAP with decoys on data 1 (MetaNovo Human SwissProt cRAP_concatenated_target_decoy) (49,076 sequences) |
|
| RAW files (Dataset Collection) | RAW files (Dataset Collection) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Identification Parameters: PAR file | toolshed.g2.bx.psu.edu/repos/galaxyp/peptideshaker/ident_params/4.0.41+galaxy1 |
| 3 | Human SwissProt (reviewed only) | toolshed.g2.bx.psu.edu/repos/galaxyp/dbbuilder/dbbuilder/0.3.4 |
| 4 | Contaminants (cRAP) | toolshed.g2.bx.psu.edu/repos/galaxyp/dbbuilder/dbbuilder/0.3.4 |
| 6 | FastaCLI__MetaNovo_Human_SwissProt_cRAP_with_decoys_on_data_1_(MetaNovo_Human_SwissProt_cRAP_concatenated_target_decoy).fasta | toolshed.g2.bx.psu.edu/repos/galaxyp/peptideshaker/fasta_cli/4.0.41+galaxy1 |
| 7 | msconvert | toolshed.g2.bx.psu.edu/repos/galaxyp/msconvert/msconvert/3.0.20287.2 |
| 8 | Human SwissProt+cRAP | toolshed.g2.bx.psu.edu/repos/galaxyp/fasta_merge_files_and_filter_unique_sequences/fasta_merge_files_and_filter_unique_sequences/1.2.0 |
| 9 | MaxQuant | toolshed.g2.bx.psu.edu/repos/galaxyp/maxquant/maxquant/2.0.3.0+galaxy0 |
| 10 | Search GUI | toolshed.g2.bx.psu.edu/repos/galaxyp/peptideshaker/search_gui/4.0.41+galaxy1 |
| 11 | FASTA-to-Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fasta_to_tabular/fasta2tab/1.1.0 |
| 12 | Select microbial peptides (MQ) | Grep1 |
| 13 | Peptide Shaker | toolshed.g2.bx.psu.edu/repos/galaxyp/peptideshaker/peptide_shaker/2.0.33+galaxy1 |
| 14 | Filter Tabular | toolshed.g2.bx.psu.edu/repos/iuc/filter_tabular/filter_tabular/3.3.0 |
| 15 | MQ Peptide Sequences | Cut1 |
| 16 | Select microbial peptides (SGPS) | Grep1 |
| 17 | Select microbial PSMs | Grep1 |
| 18 | Remove beginning | Remove beginning1 |
| 19 | Filter confident microbial peptides | Filter1 |
| 20 | Filter confident microbial PSMs | Filter1 |
| 21 | MQ Distinct Peptides | Grouping1 |
| 22 | Query Tabular | toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.0 |
| 23 | Cut | Cut1 |
| 24 | SGPS Distinct Peptides | Grouping1 |
| 25 | SGPS-MQ Peptides | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/0.1.1 |
| 26 | Distinct Peptides | Grouping1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| Human SwissProt (reviewed only) | Human SwissProt (reviewed only) | n/a |
|
| Human SwissProt+cRAP | Human SwissProt+cRAP | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| Select microbial peptides (MQ) | Select microbial peptides (MQ) | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| Select microbial peptides (SGPS) | Select microbial peptides (SGPS) | n/a |
|
| Filter confident microbial peptides | Filter confident microbial peptides | n/a |
|
| Filter confident microbial PSMs | Filter confident microbial PSMs | n/a |
|
| MQ Peptides | MQ Peptides | n/a |
|
| SGPS Distinct Peptides | SGPS Distinct Peptides | n/a |
|
| SGPS-MQ Peptides | SGPS-MQ Peptides | n/a |
|
| Distinct Peptides | Distinct Peptides | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1228 Downloads: 188 Runs: 0
Created: 2nd Jun 2025 at 10:53
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master