Workflow Type: Galaxy
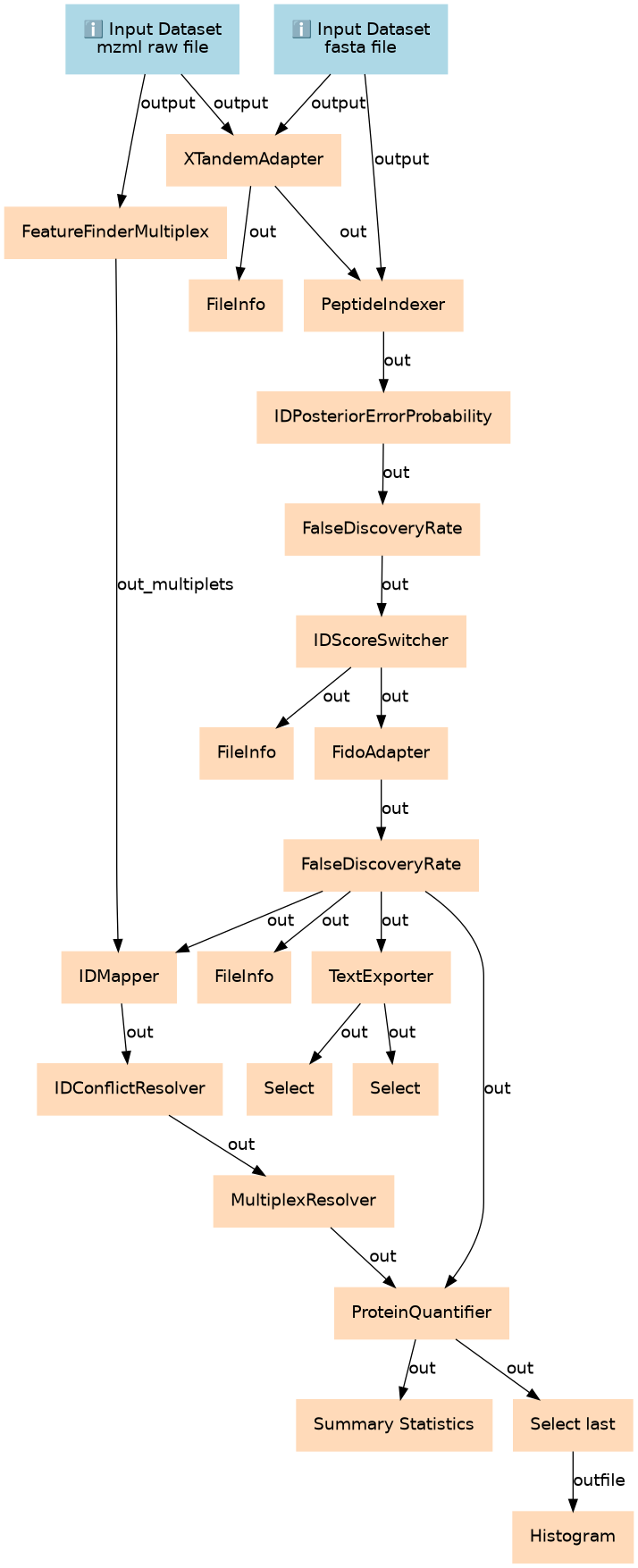
Peptide and Protein Quantification via Stable Isotope Labelling (SIL)
Associated Tutorial
This workflows is part of the tutorial Peptide and Protein Quantification via Stable Isotope Labelling (SIL), available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Florian Christoph Sigloch, Björn Grüning, Matthias Fahrner
Tutorial Contributor(s): Saskia Hiltemann, Björn Grüning, Nicola Soranzo, Helena Rasche, Melanie Föll, Bérénice Batut, Florian Christoph Sigloch, William Durand, Niall Beard, Martin Čech, Armin Dadras
Funder(s): ELIXIR Europe, de.NBI, University of Freiburg
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| fasta file | #main/fasta file | n/a |
|
| mzml raw file | #main/mzml raw file | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | FeatureFinderMultiplex | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_featurefindermultiplex/FeatureFinderMultiplex/2.6+galaxy0 |
| 3 | XTandemAdapter | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_xtandemadapter/XTandemAdapter/2.6+galaxy0 |
| 4 | FileInfo | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_fileinfo/FileInfo/2.6+galaxy0 |
| 5 | PeptideIndexer | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_peptideindexer/PeptideIndexer/2.6+galaxy0 |
| 6 | IDPosteriorErrorProbability | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_idposteriorerrorprobability/IDPosteriorErrorProbability/2.6+galaxy0 |
| 7 | FalseDiscoveryRate | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_falsediscoveryrate/FalseDiscoveryRate/2.6+galaxy0 |
| 8 | IDScoreSwitcher | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_idscoreswitcher/IDScoreSwitcher/2.6+galaxy0 |
| 9 | FileInfo | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_fileinfo/FileInfo/2.6+galaxy0 |
| 10 | FidoAdapter | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_fidoadapter/FidoAdapter/2.6+galaxy0 |
| 11 | FalseDiscoveryRate | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_falsediscoveryrate/FalseDiscoveryRate/2.6+galaxy0 |
| 12 | FileInfo | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_fileinfo/FileInfo/2.6+galaxy0 |
| 13 | TextExporter | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_textexporter/TextExporter/2.6+galaxy0 |
| 14 | IDMapper | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_idmapper/IDMapper/2.6+galaxy0 |
| 15 | Select | Grep1 |
| 16 | Select | Grep1 |
| 17 | IDConflictResolver | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_idconflictresolver/IDConflictResolver/2.6+galaxy0 |
| 18 | MultiplexResolver | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_multiplexresolver/MultiplexResolver/2.5+galaxy0 |
| 19 | ProteinQuantifier | toolshed.g2.bx.psu.edu/repos/galaxyp/openms_proteinquantifier/ProteinQuantifier/2.6+galaxy0 |
| 20 | Summary Statistics | Summary_Statistics1 |
| 21 | Select last | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_tail_tool/1.1.0 |
| 22 | Histogram | toolshed.g2.bx.psu.edu/repos/devteam/histogram/histogram_rpy/1.0.4 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_22 | #main/_anonymous_output_22 | n/a |
|
| _anonymous_output_23 | #main/_anonymous_output_23 | n/a |
|
| _anonymous_output_24 | #main/_anonymous_output_24 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1295 Downloads: 184 Runs: 0
Created: 2nd Jun 2025 at 10:53
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master