Workflow Type: Galaxy
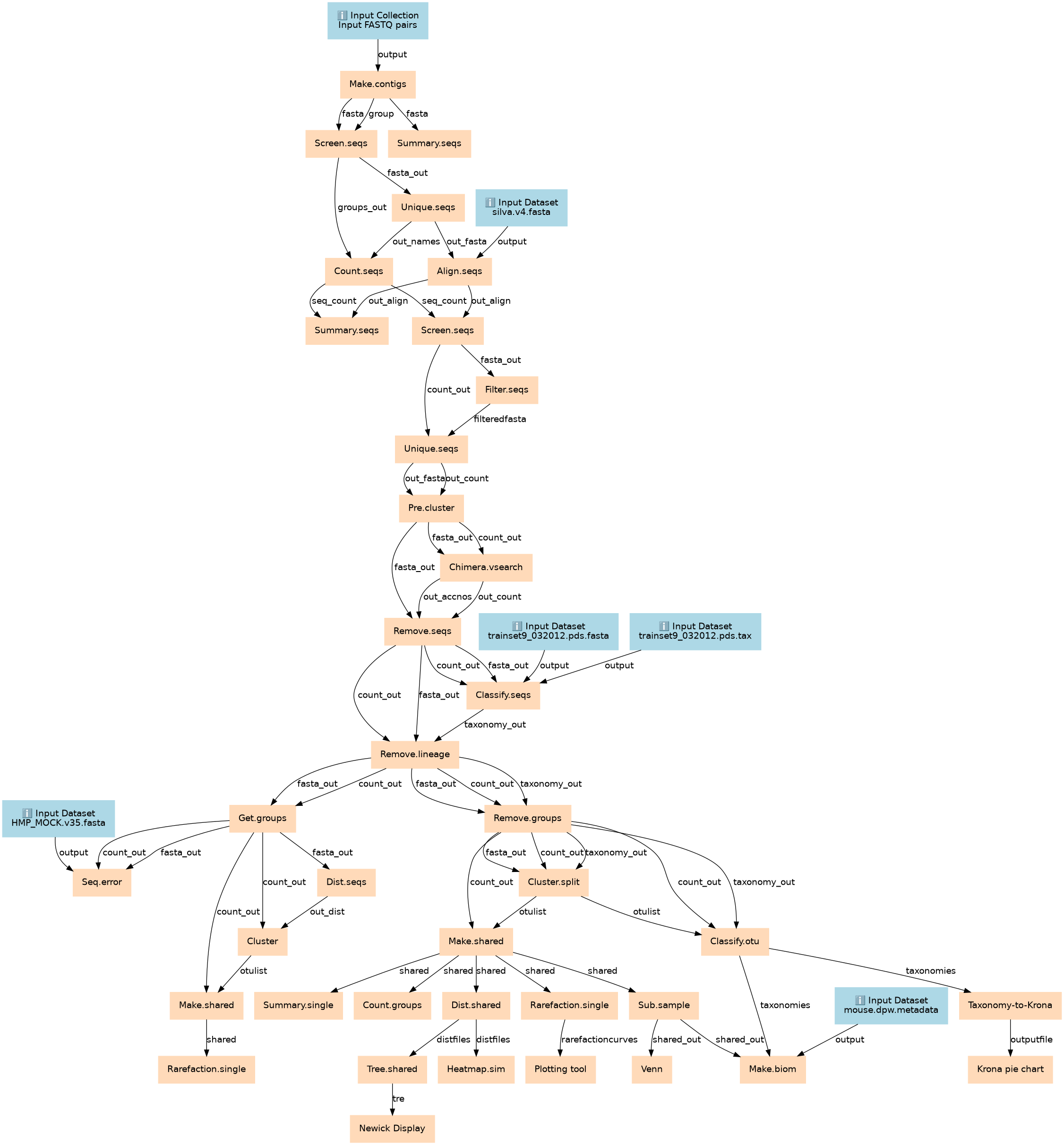
16S Microbial Analysis with mothur (extended)
Associated Tutorial
This workflows is part of the tutorial 16S Microbial Analysis with mothur (extended), available in the GTN
Thanks to...
Tutorial Author(s): Saskia Hiltemann, Bérénice Batut, Dave Clements
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| HMP_MOCK.v35.fasta | HMP_MOCK.v35.fasta | n/a |
|
| Input FASTQ pairs | Input FASTQ pairs | n/a |
|
| mouse.dpw.metadata | mouse.dpw.metadata | n/a |
|
| silva.v4.fasta | silva.v4.fasta | n/a |
|
| trainset9_032012.pds.fasta | trainset9_032012.pds.fasta | n/a |
|
| trainset9_032012.pds.tax | trainset9_032012.pds.tax | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 10 | Count.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_count_seqs/mothur_count_seqs/1.39.5.0 |
| 11 | Align.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_align_seqs/mothur_align_seqs/1.39.5.0 |
| 12 | Summary.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_summary_seqs/mothur_summary_seqs/1.39.5.0 |
| 13 | Screen.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_screen_seqs/mothur_screen_seqs/1.39.5.0 |
| 14 | Filter.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_filter_seqs/mothur_filter_seqs/1.39.5.0 |
| 15 | Unique.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_unique_seqs/mothur_unique_seqs/1.39.5.0 |
| 16 | Pre.cluster | toolshed.g2.bx.psu.edu/repos/iuc/mothur_pre_cluster/mothur_pre_cluster/1.39.5.0 |
| 17 | Chimera.vsearch | toolshed.g2.bx.psu.edu/repos/iuc/mothur_chimera_vsearch/mothur_chimera_vsearch/1.39.5.1 |
| 18 | Remove.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_remove_seqs/mothur_remove_seqs/1.36.1.0 |
| 19 | Classify.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_classify_seqs/mothur_classify_seqs/1.39.5.0 |
| 20 | Remove.lineage | toolshed.g2.bx.psu.edu/repos/iuc/mothur_remove_lineage/mothur_remove_lineage/1.39.5.0 |
| 21 | Get.groups | toolshed.g2.bx.psu.edu/repos/iuc/mothur_get_groups/mothur_get_groups/1.39.5.0 |
| 22 | Remove.groups | toolshed.g2.bx.psu.edu/repos/iuc/mothur_remove_groups/mothur_remove_groups/1.39.5.0 |
| 23 | Seq.error | toolshed.g2.bx.psu.edu/repos/iuc/mothur_seq_error/mothur_seq_error/1.39.5.0 |
| 24 | Dist.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_dist_seqs/mothur_dist_seqs/1.39.5.0 |
| 25 | Cluster.split | toolshed.g2.bx.psu.edu/repos/iuc/mothur_cluster_split/mothur_cluster_split/1.39.5.0 |
| 26 | Cluster | toolshed.g2.bx.psu.edu/repos/iuc/mothur_cluster/mothur_cluster/1.39.5.0 |
| 27 | Make.shared | toolshed.g2.bx.psu.edu/repos/iuc/mothur_make_shared/mothur_make_shared/1.39.5.0 |
| 28 | Classify.otu | toolshed.g2.bx.psu.edu/repos/iuc/mothur_classify_otu/mothur_classify_otu/1.39.5.0 |
| 29 | Make.shared | toolshed.g2.bx.psu.edu/repos/iuc/mothur_make_shared/mothur_make_shared/1.39.5.0 |
| 30 | Summary.single | toolshed.g2.bx.psu.edu/repos/iuc/mothur_summary_single/mothur_summary_single/1.39.5.0 |
| 31 | Count.groups | toolshed.g2.bx.psu.edu/repos/iuc/mothur_count_groups/mothur_count_groups/1.39.5.0 |
| 32 | Dist.shared | toolshed.g2.bx.psu.edu/repos/iuc/mothur_dist_shared/mothur_dist_shared/1.39.5.0 |
| 33 | Rarefaction.single | toolshed.g2.bx.psu.edu/repos/iuc/mothur_rarefaction_single/mothur_rarefaction_single/1.39.5.0 |
| 34 | Sub.sample | toolshed.g2.bx.psu.edu/repos/iuc/mothur_sub_sample/mothur_sub_sample/1.39.5.0 |
| 35 | Taxonomy-to-Krona | toolshed.g2.bx.psu.edu/repos/iuc/mothur_taxonomy_to_krona/mothur_taxonomy_to_krona/1.0 |
| 36 | Rarefaction.single | toolshed.g2.bx.psu.edu/repos/iuc/mothur_rarefaction_single/mothur_rarefaction_single/1.39.5.0 |
| 37 | Tree.shared | toolshed.g2.bx.psu.edu/repos/iuc/mothur_tree_shared/mothur_tree_shared/1.39.5.0 |
| 38 | Heatmap.sim | toolshed.g2.bx.psu.edu/repos/iuc/mothur_heatmap_sim/mothur_heatmap_sim/1.39.5.0 |
| 39 | Plotting tool | toolshed.g2.bx.psu.edu/repos/devteam/xy_plot/XY_Plot_1/1.0.2 |
| 40 | Venn | toolshed.g2.bx.psu.edu/repos/iuc/mothur_venn/mothur_venn/1.39.5.0 |
| 41 | Make.biom | toolshed.g2.bx.psu.edu/repos/iuc/mothur_make_biom/mothur_make_biom/1.39.5.0 |
| 42 | Krona pie chart | toolshed.g2.bx.psu.edu/repos/crs4/taxonomy_krona_chart/taxonomy_krona_chart/2.6.1.1 |
| 43 | Newick Display | toolshed.g2.bx.psu.edu/repos/iuc/newick_utils/newick_display/1.6 |
| 6 | Make.contigs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_make_contigs/mothur_make_contigs/1.39.5.0 |
| 7 | Summary.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_summary_seqs/mothur_summary_seqs/1.39.5.0 |
| 8 | Screen.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_screen_seqs/mothur_screen_seqs/1.39.5.0 |
| 9 | Unique.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_unique_seqs/mothur_unique_seqs/1.39.5.0 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1466 Downloads: 188 Runs: 0
Created: 2nd Jun 2025 at 10:54
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master