Workflow Type: Galaxy
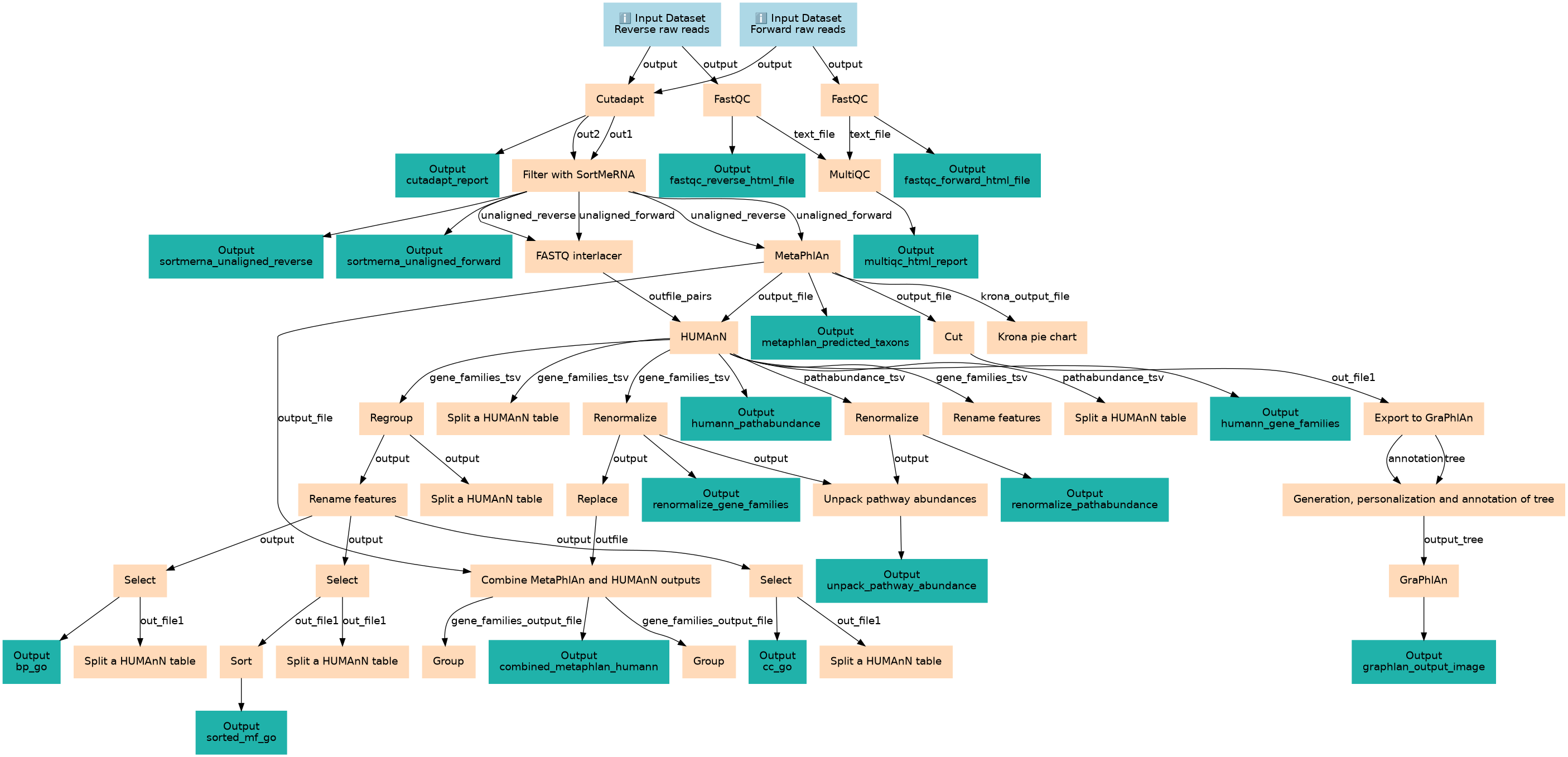
Metatranscriptomics analysis using microbiome RNA-seq data
Associated Tutorial
This workflows is part of the tutorial Metatranscriptomics analysis using microbiome RNA-seq data, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Bérénice Batut, Pratik Jagtap, Subina Mehta, Saskia Hiltemann, Paul Zierep
Tutorial Author(s): Pratik Jagtap, Subina Mehta, Ray Sajulga, Bérénice Batut, Emma Leith, Praveen Kumar, Saskia Hiltemann
Tutorial Contributor(s): Paul Zierep, Engy Nasr, Christine Oger, Saskia Hiltemann, Bérénice Batut, Helena Rasche, Björn Grüning
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Forward raw reads | Forward raw reads | Metatranscriptomics forward raw reads |
|
| Reverse raw reads | Reverse raw reads | Metatranscriptomics reverse raw reads |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 3 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 4 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/5.0+galaxy0 |
| 5 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.27+galaxy3 |
| 6 | Filter with SortMeRNA | toolshed.g2.bx.psu.edu/repos/rnateam/sortmerna/bg_sortmerna/4.3.6+galaxy0 |
| 7 | FASTQ interlacer | toolshed.g2.bx.psu.edu/repos/devteam/fastq_paired_end_interlacer/fastq_paired_end_interlacer/1.2.0.1+galaxy0 |
| 8 | MetaPhlAn | toolshed.g2.bx.psu.edu/repos/iuc/metaphlan/metaphlan/4.1.1+galaxy4 |
| 9 | Cut | Cut1 |
| 10 | Krona pie chart | toolshed.g2.bx.psu.edu/repos/crs4/taxonomy_krona_chart/taxonomy_krona_chart/2.7.1+galaxy0 |
| 11 | HUMAnN | toolshed.g2.bx.psu.edu/repos/iuc/humann/humann/3.9+galaxy0 |
| 12 | Export to GraPhlAn | toolshed.g2.bx.psu.edu/repos/iuc/export2graphlan/export2graphlan/0.20+galaxy0 |
| 13 | Split a HUMAnN table | toolshed.g2.bx.psu.edu/repos/iuc/humann_split_stratified_table/humann_split_stratified_table/3.9+galaxy0 |
| 14 | Regroup | toolshed.g2.bx.psu.edu/repos/iuc/humann_regroup_table/humann_regroup_table/3.9+galaxy0 |
| 15 | Renormalize | toolshed.g2.bx.psu.edu/repos/iuc/humann_renorm_table/humann_renorm_table/3.9+galaxy0 |
| 16 | Rename features | toolshed.g2.bx.psu.edu/repos/iuc/humann_rename_table/humann_rename_table/3.9+galaxy0 |
| 17 | Renormalize | toolshed.g2.bx.psu.edu/repos/iuc/humann_renorm_table/humann_renorm_table/3.9+galaxy0 |
| 18 | Split a HUMAnN table | toolshed.g2.bx.psu.edu/repos/iuc/humann_split_stratified_table/humann_split_stratified_table/3.9+galaxy0 |
| 19 | Generation, personalization and annotation of tree | toolshed.g2.bx.psu.edu/repos/iuc/graphlan_annotate/graphlan_annotate/1.1.3 |
| 20 | Split a HUMAnN table | toolshed.g2.bx.psu.edu/repos/iuc/humann_split_stratified_table/humann_split_stratified_table/3.9+galaxy0 |
| 21 | Rename features | toolshed.g2.bx.psu.edu/repos/iuc/humann_rename_table/humann_rename_table/3.9+galaxy0 |
| 22 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/9.5+galaxy2 |
| 23 | Unpack pathway abundances | toolshed.g2.bx.psu.edu/repos/iuc/humann_unpack_pathways/humann_unpack_pathways/3.9+galaxy0 |
| 24 | GraPhlAn | toolshed.g2.bx.psu.edu/repos/iuc/graphlan/graphlan/1.1.3 |
| 25 | Select | Grep1 |
| 26 | Select | Grep1 |
| 27 | Select | Grep1 |
| 28 | Combine MetaPhlAn and HUMAnN outputs | toolshed.g2.bx.psu.edu/repos/bebatut/combine_metaphlan2_humann2/combine_metaphlan_humann/0.3.0 |
| 29 | Split a HUMAnN table | toolshed.g2.bx.psu.edu/repos/iuc/humann_split_stratified_table/humann_split_stratified_table/3.9+galaxy0 |
| 30 | Split a HUMAnN table | toolshed.g2.bx.psu.edu/repos/iuc/humann_split_stratified_table/humann_split_stratified_table/3.9+galaxy0 |
| 31 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.5+galaxy2 |
| 32 | Split a HUMAnN table | toolshed.g2.bx.psu.edu/repos/iuc/humann_split_stratified_table/humann_split_stratified_table/3.9+galaxy0 |
| 33 | Group | Grouping1 |
| 34 | Group | Grouping1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| fastqc_reverse_html_file | fastqc_reverse_html_file | n/a |
|
| fastqc_forward_html_file | fastqc_forward_html_file | n/a |
|
| cutadapt_report | cutadapt_report | n/a |
|
| multiqc_html_report | multiqc_html_report | n/a |
|
| sortmerna_unaligned_reverse | sortmerna_unaligned_reverse | n/a |
|
| sortmerna_unaligned_forward | sortmerna_unaligned_forward | n/a |
|
| metaphlan_predicted_taxons | metaphlan_predicted_taxons | n/a |
|
| humann_pathabundance | humann_pathabundance | n/a |
|
| humann_gene_families | humann_gene_families | n/a |
|
| renormalize_gene_families | renormalize_gene_families | n/a |
|
| renormalize_pathabundance | renormalize_pathabundance | n/a |
|
| unpack_pathway_abundance | unpack_pathway_abundance | n/a |
|
| graphlan_output_image | graphlan_output_image | n/a |
|
| cc_go | cc_go | n/a |
|
| bp_go | bp_go | n/a |
|
| combined_metaphlan_humann | combined_metaphlan_humann | n/a |
|
| sorted_mf_go | sorted_mf_go | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
License
Activity
Views: 1626 Downloads: 311 Runs: 2
Created: 2nd Jun 2025 at 10:54
Last updated: 23rd Jun 2025 at 14:13
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master 1.0
1.0