Workflow Type: Galaxy
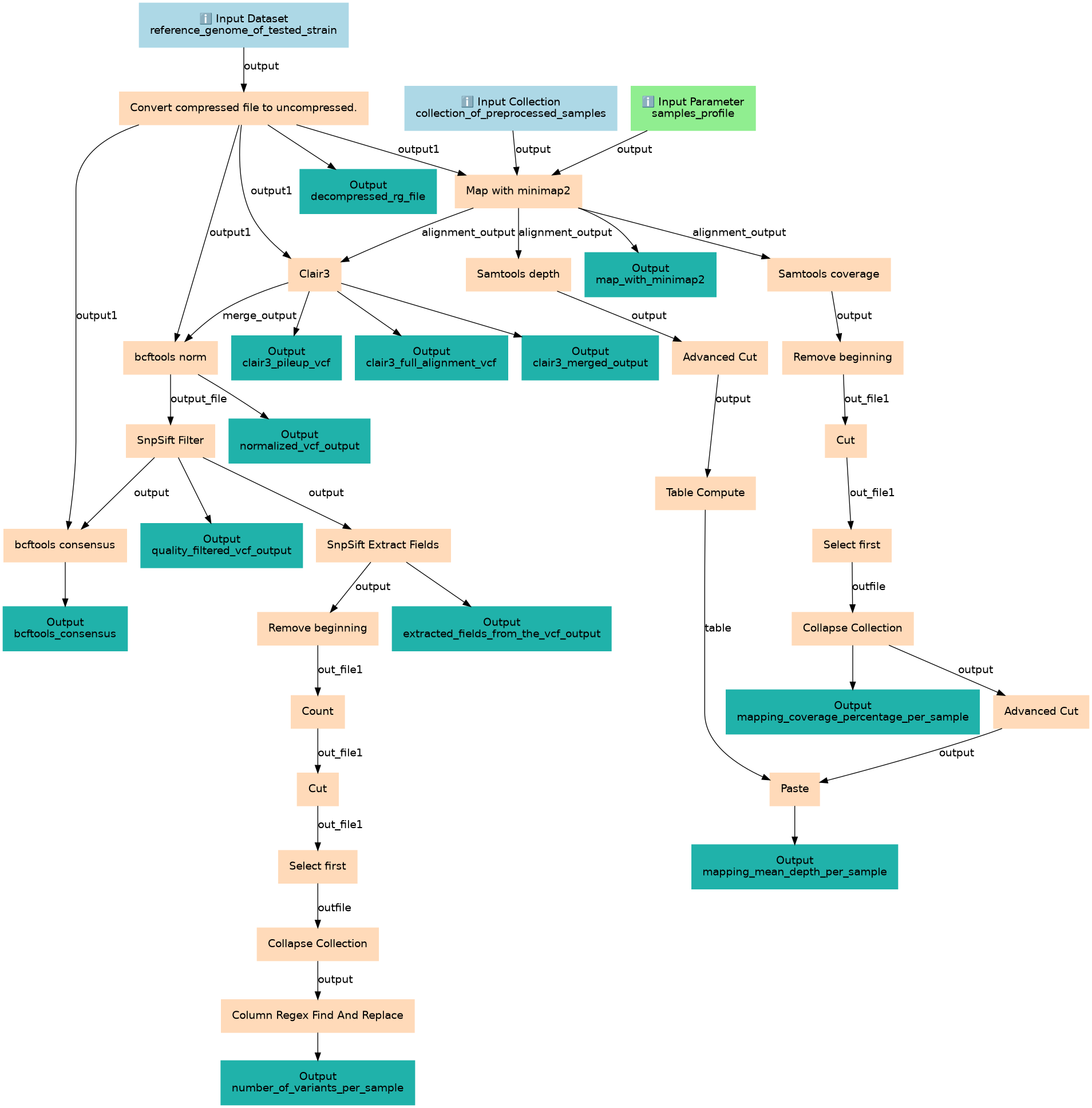
Microbiome - Variant calling and Consensus Building
Associated Tutorial
This workflows is part of the tutorial Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Engy Nasr, Bérénice Batut, Paul Zierep
Tutorial Author(s): Bérénice Batut, Engy Nasr, Paul Zierep
Tutorial Contributor(s): Hans-Rudolf Hotz, Wolfgang Maier, Saskia Hiltemann, Deepti Varshney, Paul Zierep, Bérénice Batut, Björn Grüning, Cristóbal Gallardo, Engy Nasr, Helena Rasche
Grants(s): Gallantries: Bridging Training Communities in Life Science, Environment and Health, EOSC-Life
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| collection_of_preprocessed_samples | collection_of_preprocessed_samples | Output collection from the Nanopore Preprocessing workflow |
|
| reference_genome_of_tested_strain | reference_genome_of_tested_strain | Can be built in the tool later |
|
| samples_profile | samples_profile | based on the lab preparation of the samples during sequencing, there should be a sample profile better than the other, to be chosen as an optional input to Minimap2. e.g. PacBio/Oxford Nanpore For more details check: https://github.com/lh3/minimap2?tab=readme-ov-file#use-cases |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Convert compressed file to uncompressed. | CONVERTER_gz_to_uncompressed |
| 4 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.24+galaxy0 |
| 5 | Clair3 | toolshed.g2.bx.psu.edu/repos/iuc/clair3/clair3/0.1.12+galaxy0 |
| 6 | Samtools depth | toolshed.g2.bx.psu.edu/repos/iuc/samtools_depth/samtools_depth/1.15.1+galaxy2 |
| 7 | Samtools coverage | toolshed.g2.bx.psu.edu/repos/iuc/samtools_coverage/samtools_coverage/1.15.1+galaxy2 |
| 8 | bcftools norm | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_norm/bcftools_norm/1.9+galaxy1 |
| 9 | Advanced Cut | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.3+galaxy1 |
| 10 | Remove beginning | Remove beginning1 |
| 11 | SnpSift Filter | toolshed.g2.bx.psu.edu/repos/iuc/snpsift/snpSift_filter/4.3+t.galaxy1 |
| 12 | Table Compute | toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy0 |
| 13 | Cut | Cut1 |
| 14 | SnpSift Extract Fields | toolshed.g2.bx.psu.edu/repos/iuc/snpsift/snpSift_extractFields/4.3+t.galaxy0 |
| 15 | bcftools consensus | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_consensus/bcftools_consensus/1.9+galaxy1 |
| 16 | Select first | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_head_tool/9.3+galaxy1 |
| 17 | Remove beginning | Remove beginning1 |
| 18 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 19 | Count | Count1 |
| 20 | Advanced Cut | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.3+galaxy1 |
| 21 | Cut | Cut1 |
| 22 | Paste | Paste1 |
| 23 | Select first | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_head_tool/9.3+galaxy1 |
| 24 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 25 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| decompressed_rg_file | decompressed_rg_file | n/a |
|
| map_with_minimap2 | map_with_minimap2 | n/a |
|
| clair3_pileup_vcf | clair3_pileup_vcf | n/a |
|
| clair3_full_alignment_vcf | clair3_full_alignment_vcf | n/a |
|
| clair3_merged_output | clair3_merged_output | n/a |
|
| normalized_vcf_output | normalized_vcf_output | n/a |
|
| quality_filtered_vcf_output | quality_filtered_vcf_output | n/a |
|
| extracted_fields_from_the_vcf_output | extracted_fields_from_the_vcf_output | n/a |
|
| bcftools_consensus | bcftools_consensus | n/a |
|
| mapping_coverage_percentage_per_sample | mapping_coverage_percentage_per_sample | n/a |
|
| mapping_mean_depth_per_sample | mapping_mean_depth_per_sample | n/a |
|
| number_of_variants_per_sample | number_of_variants_per_sample | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 1387 Downloads: 187 Runs: 0
Created: 2nd Jun 2025 at 10:55
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master