Workflow Type: Galaxy
Microbiome - Taxonomy Profiling
Associated Tutorial
This workflows is part of the tutorial Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Engy Nasr, Bérénice Batut, Paul Zierep
Tutorial Author(s): Bérénice Batut, Engy Nasr, Paul Zierep
Tutorial Contributor(s): Hans-Rudolf Hotz, Wolfgang Maier, Saskia Hiltemann, Deepti Varshney, Paul Zierep, Bérénice Batut, Björn Grüning, Cristóbal Gallardo, Engy Nasr, Helena Rasche
Grants(s): Gallantries: Bridging Training Communities in Life Science, Environment and Health, EOSC-Life
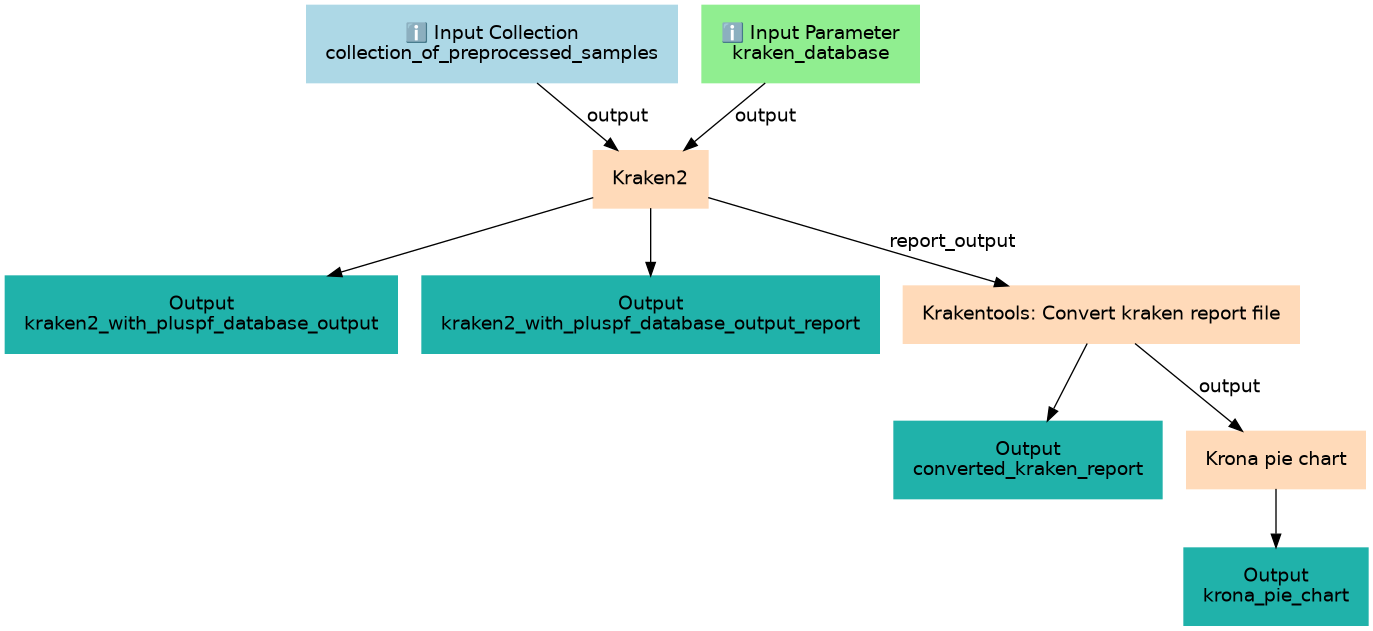
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| collection_of_preprocessed_samples | #main/collection_of_preprocessed_samples | Output collection from the Nanopore Preprocessing workflow |
|
| kraken_database | #main/kraken_database | kraken_database_for_user_to_choose |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Kraken2 | toolshed.g2.bx.psu.edu/repos/iuc/kraken2/kraken2/2.1.1+galaxy1 |
| 3 | Krakentools: Convert kraken report file | toolshed.g2.bx.psu.edu/repos/iuc/krakentools_kreport2krona/krakentools_kreport2krona/1.2+galaxy1 |
| 4 | Krona pie chart | toolshed.g2.bx.psu.edu/repos/crs4/taxonomy_krona_chart/taxonomy_krona_chart/2.7.1+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| converted_kraken_report | #main/converted_kraken_report | n/a |
|
| kraken2_with_pluspf_database_output | #main/kraken2_with_pluspf_database_output | n/a |
|
| kraken2_with_pluspf_database_output_report | #main/kraken2_with_pluspf_database_output_report | n/a |
|
| krona_pie_chart | #main/krona_pie_chart | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 1436 Downloads: 188 Runs: 1
Created: 2nd Jun 2025 at 10:55
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master