Workflow Type: Galaxy
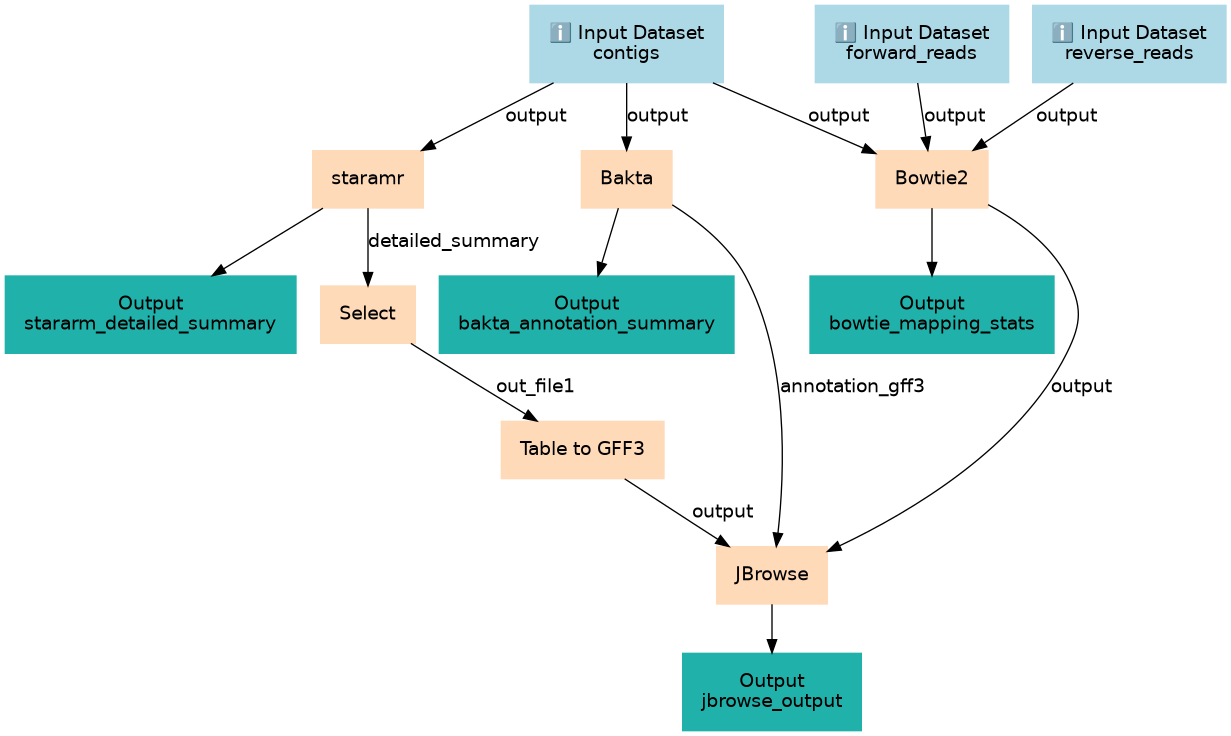
Identification of AMR genes in an assembled bacterial genome
Associated Tutorial
This workflows is part of the tutorial Identification of AMR genes in an assembled bacterial genome, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Bazante Sanders, Bérénice Batut
Tutorial Author(s): Bazante Sanders, Bérénice Batut
Tutorial Contributor(s): Helena Rasche, Bazante Sanders, Saskia Hiltemann, Miaomiao Zhou, Deepti Varshney, pimarin, Bérénice Batut, Anthony Bretaudeau, Björn Grüning
Funder(s): Avans Hogeschool, ABRomics, ELIXIR Europe
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| contigs | contigs | n/a |
|
| forward_reads | forward_reads | n/a |
|
| reverse_reads | reverse_reads | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | staramr | toolshed.g2.bx.psu.edu/repos/nml/staramr/staramr_search/0.10.0+galaxy1 |
| 4 | Bakta | toolshed.g2.bx.psu.edu/repos/iuc/bakta/bakta/1.8.2+galaxy0 |
| 5 | Bowtie2 | toolshed.g2.bx.psu.edu/repos/devteam/bowtie2/bowtie2/2.5.0+galaxy0 |
| 6 | Select | Grep1 |
| 7 | Table to GFF3 | toolshed.g2.bx.psu.edu/repos/iuc/tbl2gff3/tbl2gff3/1.2 |
| 8 | JBrowse | toolshed.g2.bx.psu.edu/repos/iuc/jbrowse/jbrowse/1.16.11+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| stararm_detailed_summary | stararm_detailed_summary | n/a |
|
| bakta_annotation_summary | bakta_annotation_summary | n/a |
|
| bowtie_mapping_stats | bowtie_mapping_stats | n/a |
|
| jbrowse_output | jbrowse_output | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 1305 Downloads: 184 Runs: 0
Created: 2nd Jun 2025 at 10:55
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master