Workflow Type: Galaxy
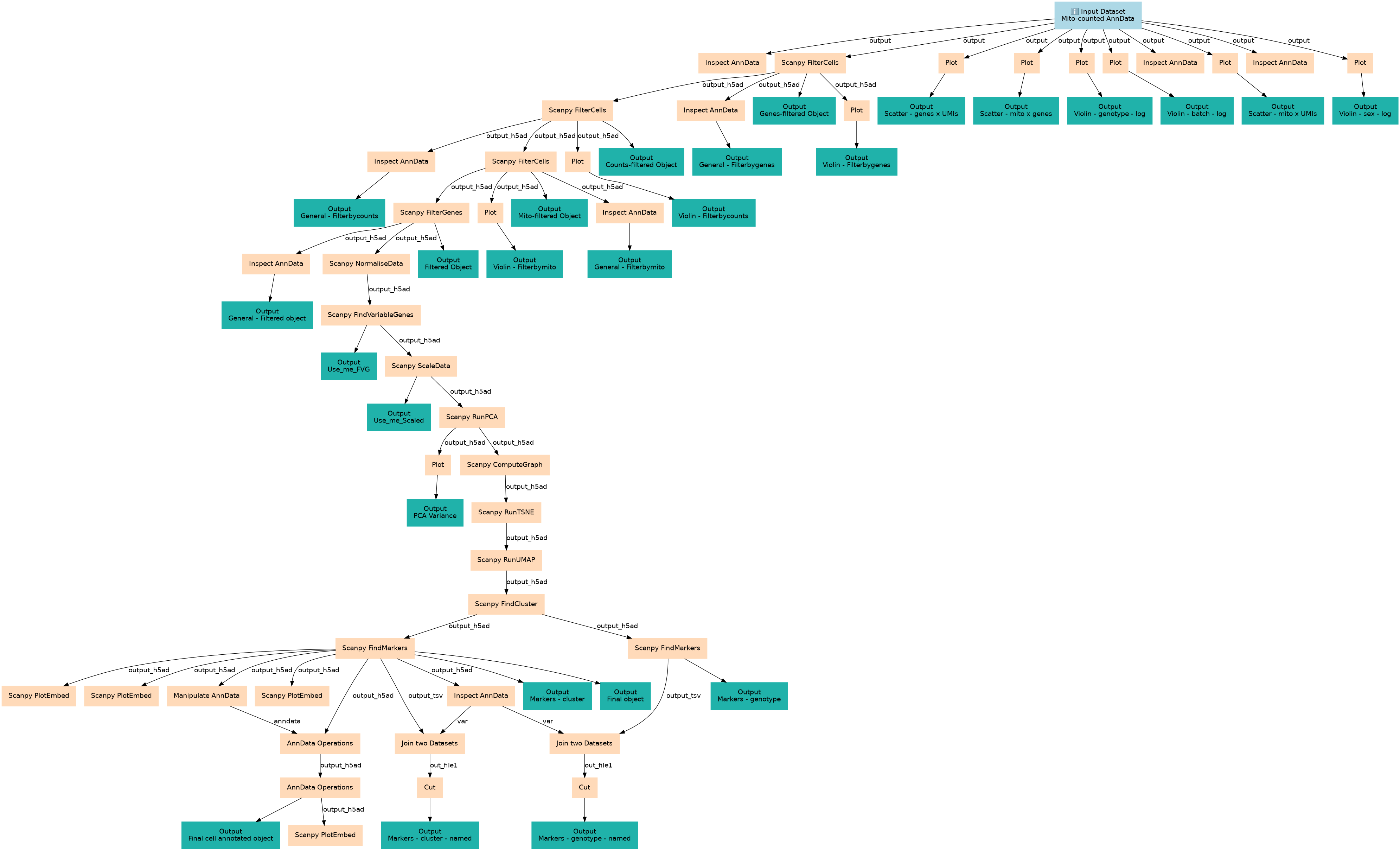
Filter, Plot and Explore Single-cell RNA-seq Data
Associated Tutorial
This workflows is part of the tutorial Filter, plot and explore single-cell RNA-seq data with Scanpy, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Wendi Bacon, Julia Jakiela
Tutorial Author(s): Wendi Bacon
Tutorial Contributor(s): Helena Rasche, Julia Jakiela, Wendi Bacon, Pavankumar Videm, Amirhossein Naghsh Nilchi, Björn Grüning, Marisa Loach, Saskia Hiltemann, Mehmet Tekman, Pablo Moreno, Matthias Bernt, Martin Čech, David López
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Mito-counted AnnData | #main/Mito-counted AnnData | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 2 | Scanpy FilterCells | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9 |
| 3 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 4 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 5 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 6 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 7 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 8 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 9 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 10 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 11 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 12 | Scanpy FilterCells | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9 |
| 13 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 14 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 15 | Scanpy FilterCells | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9 |
| 16 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 17 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 18 | Scanpy FilterGenes | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_genes/scanpy_filter_genes/1.8.1+galaxy9 |
| 19 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 20 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 21 | Scanpy NormaliseData | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_normalise_data/scanpy_normalise_data/1.8.1+galaxy9 |
| 22 | Scanpy FindVariableGenes | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_find_variable_genes/scanpy_find_variable_genes/1.8.1+galaxy9 |
| 23 | Scanpy ScaleData | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_scale_data/scanpy_scale_data/1.8.1+galaxy9 |
| 24 | Scanpy RunPCA | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_run_pca/scanpy_run_pca/1.8.1+galaxy9 |
| 25 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 26 | Scanpy ComputeGraph | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_compute_graph/scanpy_compute_graph/1.8.1+galaxy9 |
| 27 | Scanpy RunTSNE | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_run_tsne/scanpy_run_tsne/1.8.1+galaxy9 |
| 28 | Scanpy RunUMAP | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_run_umap/scanpy_run_umap/1.8.1+galaxy9 |
| 29 | Scanpy FindCluster | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_find_cluster/scanpy_find_cluster/1.8.1+galaxy9 |
| 30 | Scanpy FindMarkers | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_find_markers/scanpy_find_markers/1.8.1+galaxy9 |
| 31 | Scanpy FindMarkers | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_find_markers/scanpy_find_markers/1.8.1+galaxy9 |
| 32 | Scanpy PlotEmbed | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_plot_embed/scanpy_plot_embed/1.8.1+galaxy9 |
| 33 | Scanpy PlotEmbed | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_plot_embed/scanpy_plot_embed/1.8.1+galaxy9 |
| 34 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1 |
| 35 | Scanpy PlotEmbed | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_plot_embed/scanpy_plot_embed/1.8.1+galaxy9 |
| 36 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 37 | AnnData Operations | toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.8.1+galaxy91 |
| 38 | Join two Datasets | join1 |
| 39 | Join two Datasets | join1 |
| 40 | AnnData Operations | toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.8.1+galaxy91 |
| 41 | Cut | Cut1 |
| 42 | Cut | Cut1 |
| 43 | Scanpy PlotEmbed | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_plot_embed/scanpy_plot_embed/1.8.1+galaxy9 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Counts-filtered Object | #main/Counts-filtered Object | n/a |
|
| Filtered Object | #main/Filtered Object | n/a |
|
| Final cell annotated object | #main/Final cell annotated object | n/a |
|
| Final object | #main/Final object | n/a |
|
| General - Filterbycounts | #main/General - Filterbycounts | n/a |
|
| General - Filterbygenes | #main/General - Filterbygenes | n/a |
|
| General - Filterbymito | #main/General - Filterbymito | n/a |
|
| General - Filtered object | #main/General - Filtered object | n/a |
|
| Genes-filtered Object | #main/Genes-filtered Object | n/a |
|
| Markers - cluster | #main/Markers - cluster | n/a |
|
| Markers - cluster - named | #main/Markers - cluster - named | n/a |
|
| Markers - genotype | #main/Markers - genotype | n/a |
|
| Markers - genotype - named | #main/Markers - genotype - named | n/a |
|
| Mito-filtered Object | #main/Mito-filtered Object | n/a |
|
| PCA Variance | #main/PCA Variance | n/a |
|
| Scatter - genes x UMIs | #main/Scatter - genes x UMIs | n/a |
|
| Scatter - mito x UMIs | #main/Scatter - mito x UMIs | n/a |
|
| Scatter - mito x genes | #main/Scatter - mito x genes | n/a |
|
| Use_me_FVG | #main/Use_me_FVG | n/a |
|
| Use_me_Scaled | #main/Use_me_Scaled | n/a |
|
| Violin - Filterbycounts | #main/Violin - Filterbycounts | n/a |
|
| Violin - Filterbygenes | #main/Violin - Filterbygenes | n/a |
|
| Violin - Filterbymito | #main/Violin - Filterbymito | n/a |
|
| Violin - batch - log | #main/Violin - batch - log | n/a |
|
| Violin - genotype - log | #main/Violin - genotype - log | n/a |
|
| Violin - sex - log | #main/Violin - sex - log | n/a |
|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1320 Downloads: 190 Runs: 0
Created: 2nd Jun 2025 at 10:55
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master