Workflow Type: Galaxy
Associated Tutorial
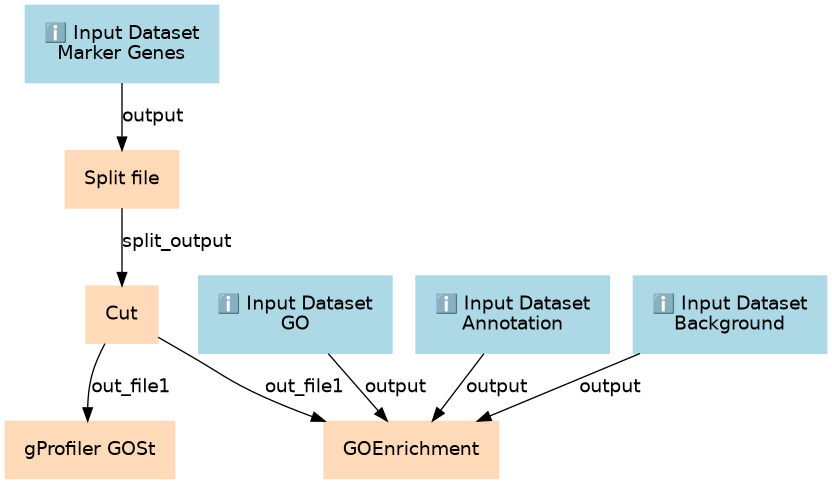
This workflows is part of the tutorial GO Enrichment Analysis on Single-Cell RNA-Seq Data, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Menna Gamal
Tutorial Author(s): Menna Gamal
Tutorial Contributor(s): Wendi Bacon, Björn Grüning, Pablo Moreno, Nicola Soranzo, Helena Rasche, Saskia Hiltemann, Mennayousef, Martin Čech, Armin Dadras
Funder(s): ELIXIR Europe, de.NBI, University of Freiburg
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Annotation | Annotation | n/a |
|
| Background | Background | n/a |
|
| GO | GO | n/a |
|
| Marker Genes | Marker Genes | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Split file | toolshed.g2.bx.psu.edu/repos/bgruening/split_file_on_column/tp_split_on_column/0.4 |
| 5 | Cut | Cut1 |
| 6 | GOEnrichment | toolshed.g2.bx.psu.edu/repos/iuc/goenrichment/goenrichment/2.0.1 |
| 7 | gProfiler GOSt | toolshed.g2.bx.psu.edu/repos/iuc/gprofiler_gost/gprofiler_gost/0.1.7+galaxy11 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 1362 Downloads: 182 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master