Workflow Type: Galaxy
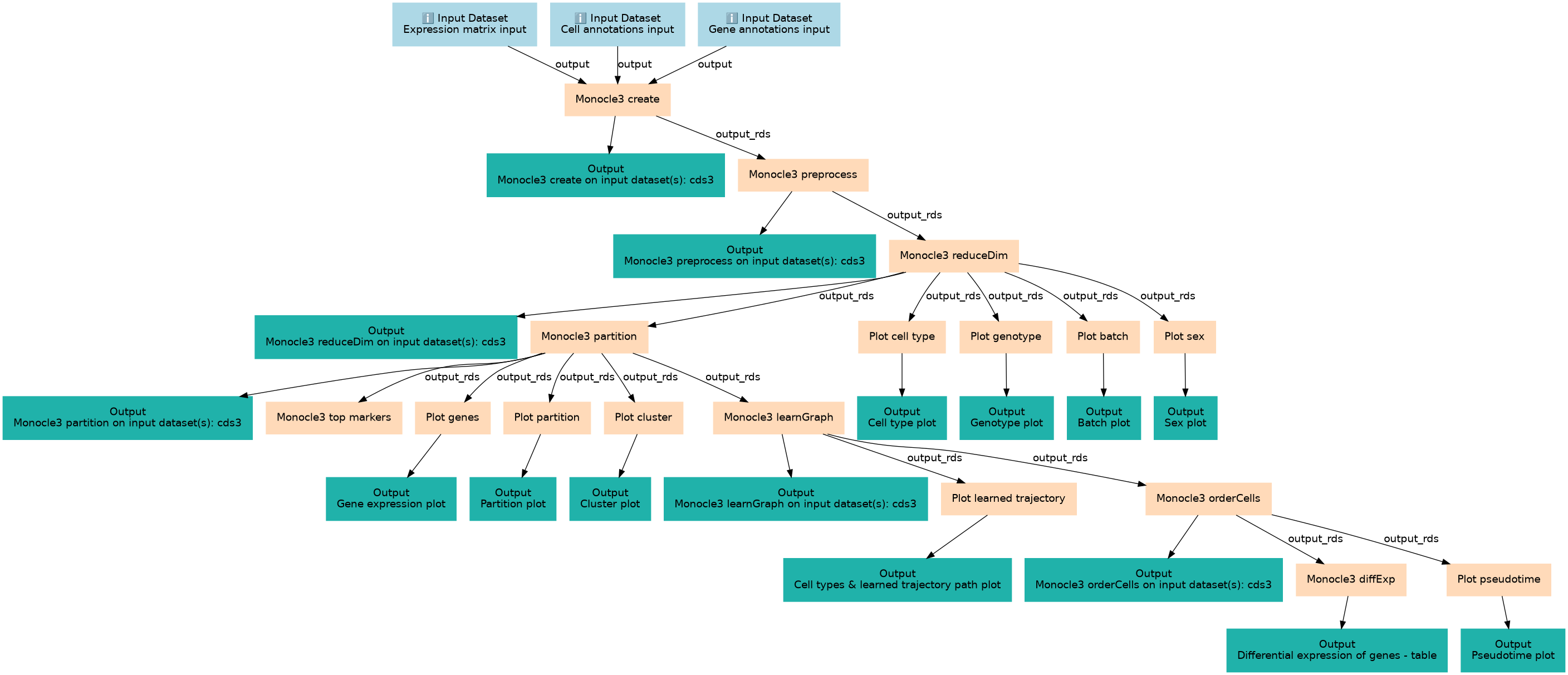
Trajectory analysis using Monocle3, starting from 3 input files: expression matrix, gene and cell annotations
Associated Tutorial
This workflows is part of the tutorial Inferring single cell trajectories with Monocle3, available in the GTN
Thanks to...
Tutorial Author(s): Julia Jakiela
Tutorial Contributor(s): Helena Rasche, Wendi Bacon, Pavankumar Videm, Julia Jakiela, Saskia Hiltemann, Mehmet Tekman, Matthias Bernt, Pablo Moreno, Björn Grüning
Grants(s): EPSRC Training Grant DTP 2020-2021 Open University
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Cell annotations input | #main/Cell annotations input | n/a |
|
| Expression matrix input | #main/Expression matrix input | n/a |
|
| Gene annotations input | #main/Gene annotations input | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Monocle3 create | Check the data format that you're using. toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_create/monocle3_create/0.1.4+galaxy2 |
| 4 | Monocle3 preprocess | You might want to change the dimensionality of the reduced space here. toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_preprocess/monocle3_preprocess/0.1.4+galaxy0 |
| 5 | Monocle3 reduceDim | UMAP/tSNE/PCA/LSI. However for further steps you need to use UMAP. toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_reducedim/monocle3_reduceDim/0.1.4+galaxy0 |
| 6 | Monocle3 partition | You might want to change resolution (affects clusters) and/or q-value (affects partitions). toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_partition/monocle3_partition/0.1.4+galaxy0 |
| 7 | Plot cell type | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 8 | Plot genotype | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 9 | Plot batch | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 10 | Plot sex | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 11 | Monocle3 top markers | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_topmarkers/monocle3_topmarkers/0.1.5+galaxy0 |
| 12 | Plot genes | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 13 | Plot partition | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 14 | Plot cluster | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 15 | Monocle3 learnGraph | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_learngraph/monocle3_learnGraph/0.1.4+galaxy0 |
| 16 | Plot learned trajectory | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
| 17 | Monocle3 orderCells | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_ordercells/monocle3_orderCells/0.1.4+galaxy0 |
| 18 | Monocle3 diffExp | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_diffexp/monocle3_diffExp/0.1.4+galaxy1 |
| 19 | Plot pseudotime | toolshed.g2.bx.psu.edu/repos/ebi-gxa/monocle3_plotcells/monocle3_plotCells/0.1.5+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Batch plot | #main/Batch plot | n/a |
|
| Cell type plot | #main/Cell type plot | n/a |
|
| Cell types & learned trajectory path plot | #main/Cell types & learned trajectory path plot | n/a |
|
| Cluster plot | #main/Cluster plot | n/a |
|
| Differential expression of genes - table | #main/Differential expression of genes - table | n/a |
|
| Gene expression plot | #main/Gene expression plot | n/a |
|
| Genotype plot | #main/Genotype plot | n/a |
|
| Monocle3 create on input dataset(s): cds3 | #main/Monocle3 create on input dataset(s): cds3 | n/a |
|
| Monocle3 learnGraph on input dataset(s): cds3 | #main/Monocle3 learnGraph on input dataset(s): cds3 | n/a |
|
| Monocle3 orderCells on input dataset(s): cds3 | #main/Monocle3 orderCells on input dataset(s): cds3 | n/a |
|
| Monocle3 partition on input dataset(s): cds3 | #main/Monocle3 partition on input dataset(s): cds3 | n/a |
|
| Monocle3 preprocess on input dataset(s): cds3 | #main/Monocle3 preprocess on input dataset(s): cds3 | n/a |
|
| Monocle3 reduceDim on input dataset(s): cds3 | #main/Monocle3 reduceDim on input dataset(s): cds3 | n/a |
|
| Partition plot | #main/Partition plot | n/a |
|
| Pseudotime plot | #main/Pseudotime plot | n/a |
|
| Sex plot | #main/Sex plot | n/a |
|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1453 Downloads: 203 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master