Workflow Type: Galaxy
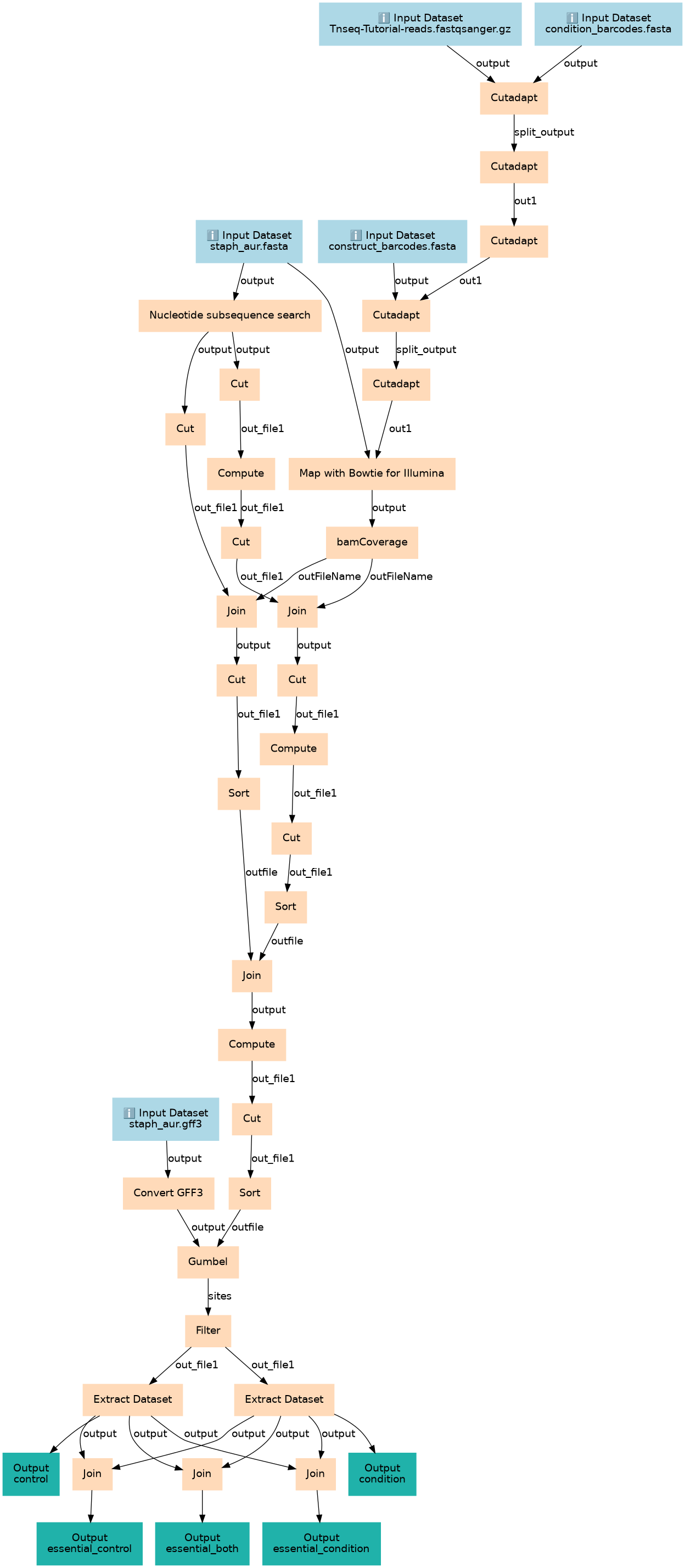
Essential genes detection with Transposon insertion sequencing
Associated Tutorial
This workflows is part of the tutorial Essential genes detection with Transposon insertion sequencing, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Delphine Lariviere, Bérénice Batut
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Tnseq-Tutorial-reads.fastqsanger.gz | #main/Tnseq-Tutorial-reads.fastqsanger.gz | n/a |
|
| condition_barcodes.fasta | #main/condition_barcodes.fasta | n/a |
|
| construct_barcodes.fasta | #main/construct_barcodes.fasta | n/a |
|
| staph_aur.fasta | #main/staph_aur.fasta | n/a |
|
| staph_aur.gff3 | #main/staph_aur.gff3 | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 5 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/1.16.5 |
| 6 | Nucleotide subsequence search | toolshed.g2.bx.psu.edu/repos/bgruening/find_subsequences/bg_find_subsequences/0.2 |
| 7 | Convert GFF3 | toolshed.g2.bx.psu.edu/repos/iuc/gff_to_prot/gff_to_prot/3.0.2+galaxy0 |
| 8 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/1.16.5 |
| 9 | Cut | Cut1 |
| 10 | Cut | Cut1 |
| 11 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/1.16.5 |
| 12 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/1.3.0 |
| 13 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/1.16.5 |
| 14 | Cut | Cut1 |
| 15 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/1.16.5 |
| 16 | Map with Bowtie for Illumina | toolshed.g2.bx.psu.edu/repos/devteam/bowtie_wrappers/bowtie_wrapper/1.2.0 |
| 17 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.2.0.0 |
| 18 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
| 19 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
| 20 | Cut | Cut1 |
| 21 | Cut | Cut1 |
| 22 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/1.1.1 |
| 23 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/1.3.0 |
| 24 | Cut | Cut1 |
| 25 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/1.1.1 |
| 26 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
| 27 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/1.3.0 |
| 28 | Cut | Cut1 |
| 29 | Sort | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/1.1.1 |
| 30 | Gumbel | toolshed.g2.bx.psu.edu/repos/iuc/transit_gumbel/transit_gumbel/3.0.2+galaxy0 |
| 31 | Filter | Filter1 |
| 32 | Extract Dataset | __EXTRACT_DATASET__ |
| 33 | Extract Dataset | __EXTRACT_DATASET__ |
| 34 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
| 35 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
| 36 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_22 | #main/_anonymous_output_22 | n/a |
|
| _anonymous_output_23 | #main/_anonymous_output_23 | n/a |
|
| _anonymous_output_24 | #main/_anonymous_output_24 | n/a |
|
| _anonymous_output_25 | #main/_anonymous_output_25 | n/a |
|
| _anonymous_output_26 | #main/_anonymous_output_26 | n/a |
|
| _anonymous_output_27 | #main/_anonymous_output_27 | n/a |
|
| _anonymous_output_28 | #main/_anonymous_output_28 | n/a |
|
| _anonymous_output_29 | #main/_anonymous_output_29 | n/a |
|
| _anonymous_output_30 | #main/_anonymous_output_30 | n/a |
|
| _anonymous_output_31 | #main/_anonymous_output_31 | n/a |
|
| _anonymous_output_32 | #main/_anonymous_output_32 | n/a |
|
| _anonymous_output_33 | #main/_anonymous_output_33 | n/a |
|
| _anonymous_output_34 | #main/_anonymous_output_34 | n/a |
|
| _anonymous_output_35 | #main/_anonymous_output_35 | n/a |
|
| _anonymous_output_36 | #main/_anonymous_output_36 | n/a |
|
| _anonymous_output_37 | #main/_anonymous_output_37 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
| condition | #main/condition | n/a |
|
| control | #main/control | n/a |
|
| essential_both | #main/essential_both | n/a |
|
| essential_condition | #main/essential_condition | n/a |
|
| essential_control | #main/essential_control | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1393 Downloads: 181 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master