Workflow Type: Galaxy
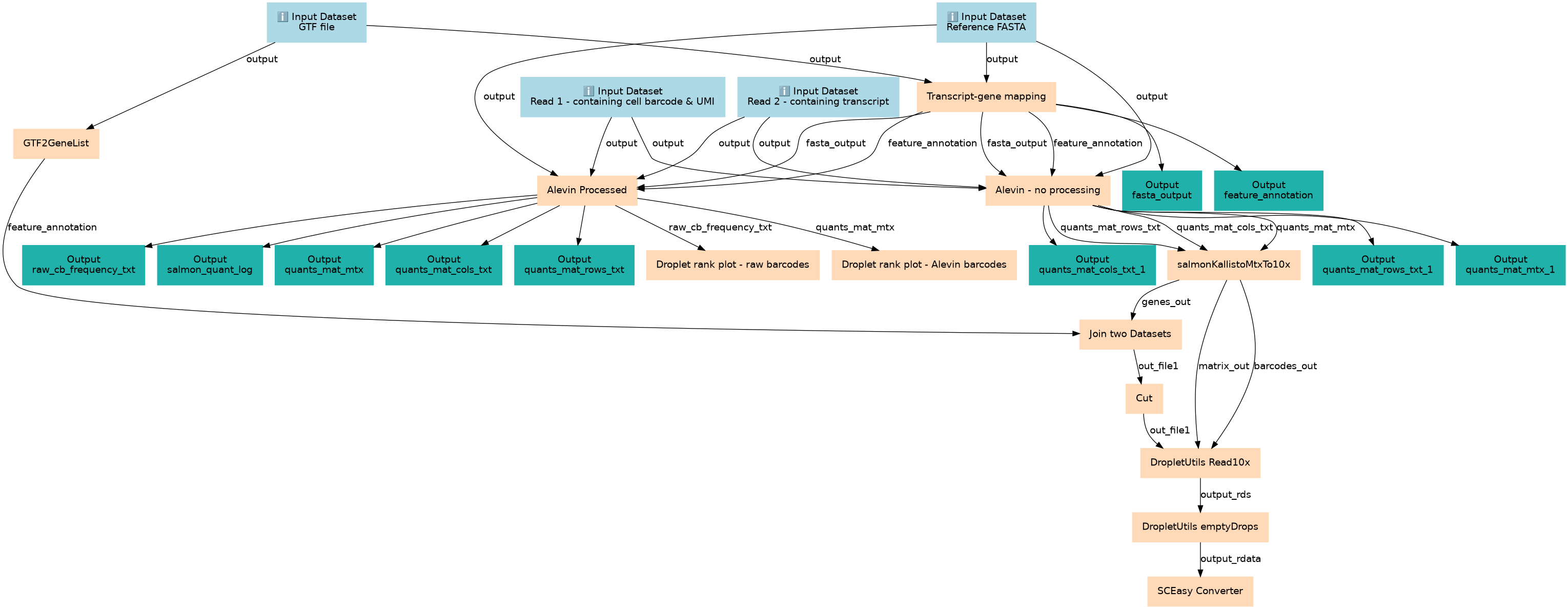
This workflow generates a single cell matrix using Alevin.
Associated Tutorial
This workflows is part of the tutorial Generating a single cell matrix using Alevin, available in the GTN
Thanks to...
Workflow Author(s): Julia Jakiela, Wendi Bacon
Tutorial Author(s): Wendi Bacon, Jonathan Manning
Tutorial Contributor(s): Helena Rasche, Julia Jakiela, Pavankumar Videm, Björn Grüning, Marisa Loach, Wendi Bacon, Teresa Müller, Saskia Hiltemann, Mehmet Tekman, Jonathan Manning, Beatriz Serrano-Solano, David López
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| GTF file | GTF file | You can find the file containing the gene information for your species here: http://www.ensembl.org/info/data/ftp/index.html/ |
|
| Read 1 - containing cell barcode & UMI | Read 1 - containing cell barcode & UMI | This should be a fast file containing the shorter read - i.e. the cell barcode and the unique molecular identifier |
|
| Read 2 - containing transcript | Read 2 - containing transcript | This should be the read containing the transcript |
|
| Reference FASTA | Reference FASTA | You can find this file for your species here: http://www.ensembl.org/info/data/ftp/index.html/ |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | GTF2GeneList | toolshed.g2.bx.psu.edu/repos/ebi-gxa/gtf2gene_list/_ensembl_gtf2gene_list/1.52.0+galaxy0 |
| 5 | Transcript-gene mapping | toolshed.g2.bx.psu.edu/repos/ebi-gxa/gtf2gene_list/_ensembl_gtf2gene_list/1.52.0+galaxy0 |
| 6 | Alevin Processed | toolshed.g2.bx.psu.edu/repos/bgruening/alevin/alevin/1.10.1+galaxy0 |
| 7 | Alevin - no processing | toolshed.g2.bx.psu.edu/repos/bgruening/alevin/alevin/1.10.1+galaxy0 |
| 8 | Droplet rank plot - raw barcodes | toolshed.g2.bx.psu.edu/repos/ebi-gxa/droplet_barcode_plot/_dropletBarcodePlot/1.6.1+galaxy2 |
| 9 | Droplet rank plot - Alevin barcodes | toolshed.g2.bx.psu.edu/repos/ebi-gxa/droplet_barcode_plot/_dropletBarcodePlot/1.6.1+galaxy2 |
| 10 | salmonKallistoMtxTo10x | toolshed.g2.bx.psu.edu/repos/ebi-gxa/salmon_kallisto_mtx_to_10x/_salmon_kallisto_mtx_to_10x/0.0.1+galaxy6 |
| 11 | Join two Datasets | join1 |
| 12 | Cut | Cut1 |
| 13 | DropletUtils Read10x | toolshed.g2.bx.psu.edu/repos/ebi-gxa/dropletutils_read_10x/dropletutils_read_10x/1.0.4+galaxy0 |
| 14 | DropletUtils emptyDrops | toolshed.g2.bx.psu.edu/repos/ebi-gxa/dropletutils_empty_drops/dropletutils_empty_drops/1.0.4+galaxy0 |
| 15 | SCEasy Converter | toolshed.g2.bx.psu.edu/repos/iuc/sceasy_convert/sceasy_convert/0.0.7+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| fasta_output | fasta_output | n/a |
|
| feature_annotation | feature_annotation | n/a |
|
| raw_cb_frequency_txt | raw_cb_frequency_txt | n/a |
|
| salmon_quant_log | salmon_quant_log | n/a |
|
| quants_mat_mtx | quants_mat_mtx | n/a |
|
| quants_mat_cols_txt | quants_mat_cols_txt | n/a |
|
| quants_mat_rows_txt | quants_mat_rows_txt | n/a |
|
| quants_mat_rows_txt_1 | quants_mat_rows_txt_1 | n/a |
|
| quants_mat_mtx_1 | quants_mat_mtx_1 | n/a |
|
| quants_mat_cols_txt_1 | quants_mat_cols_txt_1 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 1291 Downloads: 203 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master