Workflow Type: Galaxy
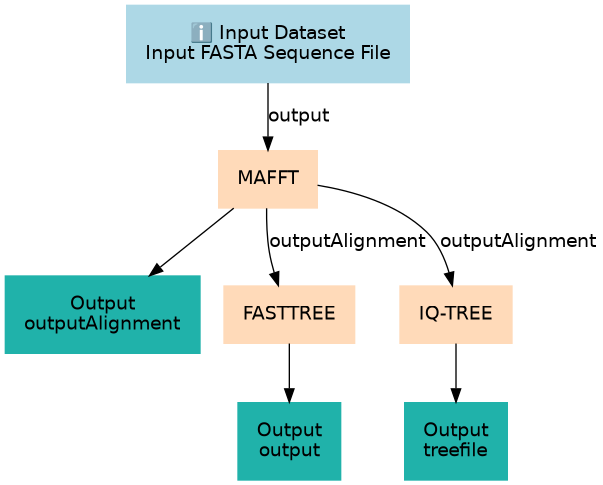
Tree building for "Phylogenetics - Back to basics"
Associated Tutorial
This workflows is part of the tutorial Phylogenetics - Back to basics, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Helena Rasche, Michael Charleston
Tutorial Author(s): Michael Charleston
Tutorial Contributor(s): Adam Taranto, Melissa Burke, Patrick Capon
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input FASTA Sequence File | #main/Input FASTA Sequence File | This should be a fasta formatted file with multiple sequences. |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | MAFFT | toolshed.g2.bx.psu.edu/repos/rnateam/mafft/rbc_mafft/7.508+galaxy1 |
| 2 | FASTTREE | toolshed.g2.bx.psu.edu/repos/iuc/fasttree/fasttree/2.1.10+galaxy1 |
| 3 | IQ-TREE | toolshed.g2.bx.psu.edu/repos/iuc/iqtree/iqtree/2.1.2+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| output | #main/output | n/a |
|
| outputAlignment | #main/outputAlignment | n/a |
|
| treefile | #main/treefile | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 1321 Downloads: 193 Runs: 0
Created: 2nd Jun 2025 at 10:58
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master