Workflow Type: Galaxy
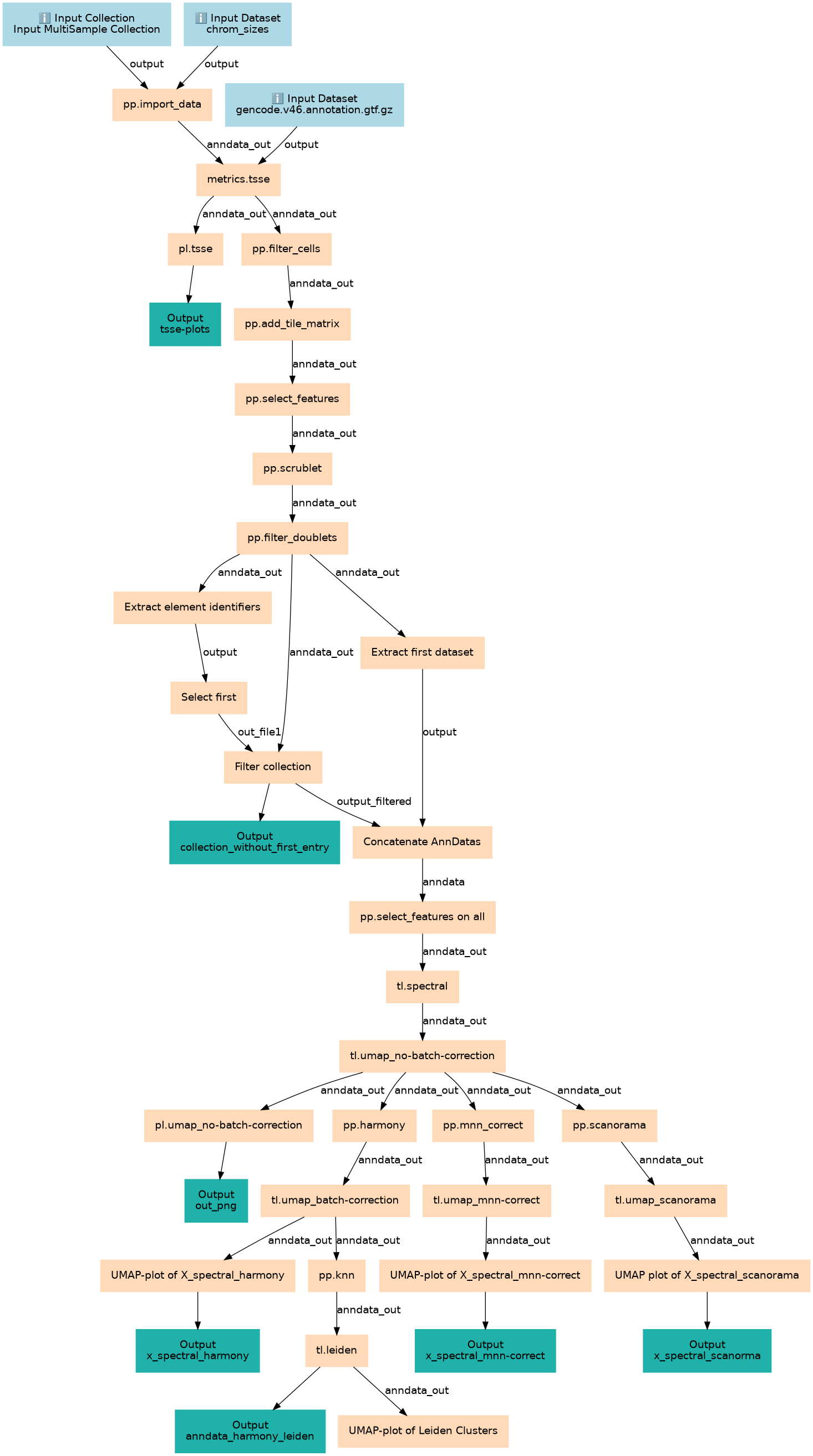
This Workflow takes a dataset collection of single-cell ATAC-seq fragments and performs:
- preprocessing
- filtering
- concatenation
- dimension reduction
- batch correction
- leiden clustering
Associated Tutorial
This workflows is part of the tutorial Multi-sample batch correction with Harmony and SnapATAC2, available in the GTN
Features
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Timon Schlegel
Tutorial Author(s): Timon Schlegel
Tutorial Contributor(s): Diana Chiang Jurado, Saskia Hiltemann, Timon Schlegel
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input MultiSample Collection | Input MultiSample Collection | n/a |
|
| chrom_sizes | chrom_sizes | n/a |
|
| gencode.v46.annotation.gtf.gz | gencode.v46.annotation.gtf.gz | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | pp.import_data | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 4 | metrics.tsse | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 5 | pl.tsse | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.6.4+galaxy1 |
| 6 | pp.filter_cells | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 7 | pp.add_tile_matrix | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 8 | pp.select_features | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 9 | pp.scrublet | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 10 | pp.filter_doublets | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 11 | Extract element identifiers | toolshed.g2.bx.psu.edu/repos/iuc/collection_element_identifiers/collection_element_identifiers/0.0.2 |
| 12 | Extract first dataset | __EXTRACT_DATASET__ |
| 13 | Select first | Show beginning1 |
| 14 | Filter collection | __FILTER_FROM_FILE__ |
| 15 | Concatenate AnnDatas | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.3+galaxy0 |
| 16 | pp.select_features on all | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 17 | tl.spectral | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 18 | tl.umap_no-batch-correction | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 19 | pl.umap_no-batch-correction | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.6.4+galaxy1 |
| 20 | pp.harmony | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 21 | pp.mnn_correct | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 22 | pp.scanorama | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_preprocessing/snapatac2_preprocessing/2.6.4+galaxy1 |
| 23 | tl.umap_batch-correction | Recalculates UMAP-embeddings and adds it under the key 'X_umap_harmony' toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 24 | tl.umap_mnn-correct | Recalculates UMAP-embeddings and adds it under the key 'X_umap_harmony' toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 25 | tl.umap_scanorama | Recalculates UMAP-embeddings and adds it under the key 'X_umap_harmony' toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 26 | UMAP-plot of X_spectral_harmony | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.6.4+galaxy1 |
| 27 | pp.knn | Recalculates UMAP-embeddings and adds it under the key 'X_umap_harmony' toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 28 | UMAP-plot of X_spectral_mnn-correct | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.6.4+galaxy1 |
| 29 | UMAP plot of X_spectral_scanorama | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.6.4+galaxy1 |
| 30 | tl.leiden | Recalculates UMAP-embeddings and adds it under the key 'X_umap_harmony' toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_clustering/snapatac2_clustering/2.6.4+galaxy1 |
| 31 | UMAP-plot of Leiden Clusters | toolshed.g2.bx.psu.edu/repos/iuc/snapatac2_plotting/snapatac2_plotting/2.6.4+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| tsse-plots | tsse-plots | n/a |
|
| collection_without_first_entry | collection_without_first_entry | n/a |
|
| out_png | out_png | n/a |
|
| x_spectral_harmony | x_spectral_harmony | n/a |
|
| x_spectral_mnn-correct | x_spectral_mnn-correct | n/a |
|
| x_spectral_scanorma | x_spectral_scanorma | n/a |
|
| anndata_harmony_leiden | anndata_harmony_leiden | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1252 Downloads: 168 Runs: 0
Created: 2nd Jun 2025 at 10:58
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master