Workflow Type: Galaxy
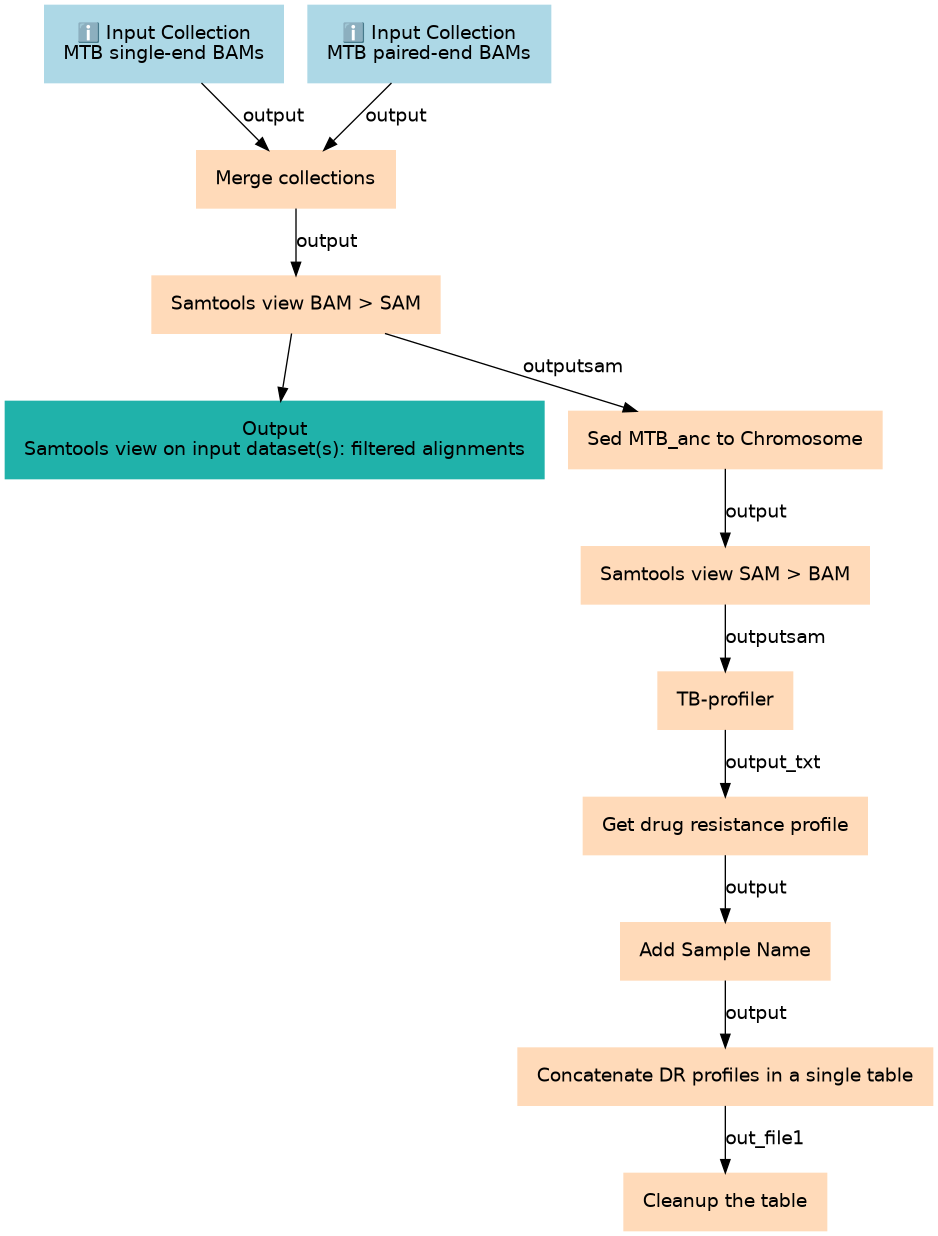
Starting from the BAM files produced by snippy, generate a table that summarizes the drug-resistance profile for each sample
Associated Tutorial
This workflows is part of the tutorial Identifying tuberculosis transmission links: from SNPs to transmission clusters, available in the GTN
Thanks to...
Tutorial Author(s): Galo A. Goig, Daniela Brites, Christoph Stritt
Tutorial Contributor(s): Wolfgang Maier, Saskia Hiltemann, Helena Rasche, Galo A. Goig, Björn Grüning, Peter van Heusden, Christoph Stritt, Lucille Delisle
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| MTB paired-end BAMs | #main/MTB paired-end BAMs | BAMs obtained from paired-end mappings with snippy |
|
| MTB single-end BAMs | #main/MTB single-end BAMs | BAMs obtained from single-end mappings with snippy |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Merge collections | Merge single-end and paired-end BAMs in a single collection to be analyzed alltogether __MERGE_COLLECTION__ |
| 3 | Samtools view BAM > SAM | We want a text (SAM) file to substitute the string "MTB_anc" by "Chromosome" so it is compatible with TB-profiler toolshed.g2.bx.psu.edu/repos/iuc/samtools_view/samtools_view/1.13+galaxy2 |
| 4 | Sed (MTB_anc to Chromosome) | Change the string MTB_anc to "Chromosome" so it is compatible with TB-profiler toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sed_tool/1.1.1 |
| 5 | Samtools view SAM > BAM | toolshed.g2.bx.psu.edu/repos/iuc/samtools_view/samtools_view/1.13+galaxy2 |
| 6 | TB-profiler | Generate TB-profiler reports with drug resistance determinants toolshed.g2.bx.psu.edu/repos/iuc/tbprofiler/tb_profiler_profile/3.0.8+galaxy0 |
| 7 | Get drug resistance profile | From TB profiler, search with `grep` the part of the text that describes the DR profile (e.g Drug-Resistance: MDR) toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/1.1.1 |
| 8 | Add Sample Name | We have generated one file per sample, that contains the DR profile (e.g Drug-Resistance: MDR) We want to prepend the name of the sample so we have: (Sample_name Drug-Resistance: MDR) toolshed.g2.bx.psu.edu/repos/mvdbeek/add_input_name_as_column/addName/0.2.0 |
| 9 | Concatenate DR profiles in a single table | The output will be: Sample_A DR_profile_A Sample_B DR_profile_B Sample_Z DR_profile_Z toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/0.1.1 |
| 10 | Cleanup the table | Remove unnecessary text from the table like ".txt" or "Drug-resistance:" toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_line/1.1.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Samtools view on input dataset(s): filtered alignments | #main/Samtools view on input dataset(s): filtered alignments | n/a |
|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 1401 Downloads: 188 Runs: 0
Created: 2nd Jun 2025 at 10:58
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master