Workflow Type: Galaxy
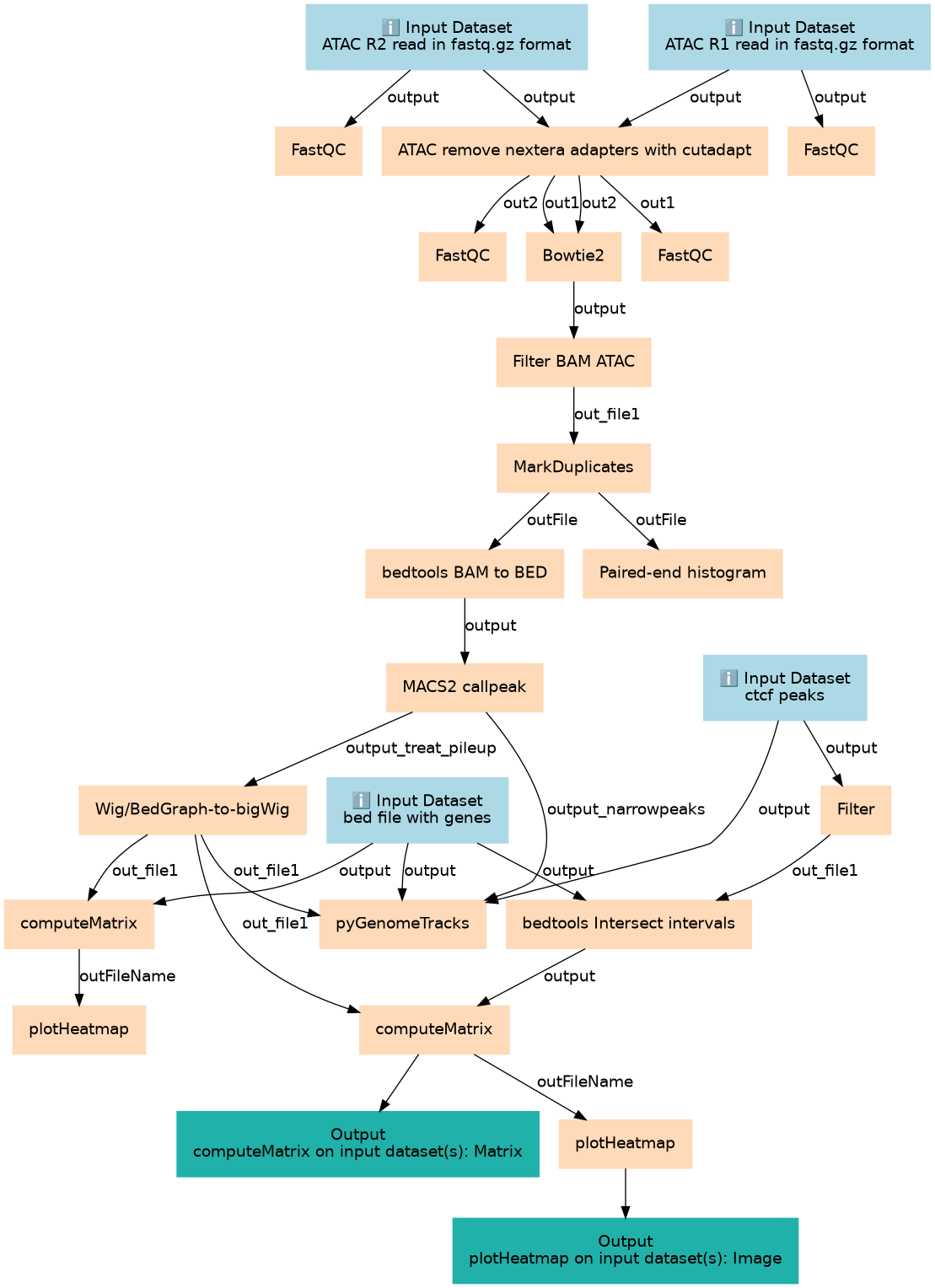
atac-seq
Associated Tutorial
This workflows is part of the tutorial ATAC-Seq data analysis, available in the GTN
Thanks to...
Tutorial Author(s): Lucille Delisle, Maria Doyle, Florian Heyl
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| ATAC R1 read in fastq(.gz) format | #main/ATAC R1 read in fastq(.gz) format | n/a |
|
| ATAC R2 read in fastq(.gz) format | #main/ATAC R2 read in fastq(.gz) format | n/a |
|
| bed file with genes | #main/bed file with genes | n/a |
|
| ctcf peaks | #main/ctcf peaks | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Filter | Filter1 |
| 5 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 6 | ATAC remove nextera adapters with cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/1.16.5 |
| 7 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 8 | bedtools Intersect intervals | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_intersectbed/2.30.0 |
| 9 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 10 | Bowtie2 | toolshed.g2.bx.psu.edu/repos/devteam/bowtie2/bowtie2/2.4.2+galaxy0 |
| 11 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 12 | Filter BAM ATAC | toolshed.g2.bx.psu.edu/repos/devteam/bamtools_filter/bamFilter/2.4.1 |
| 13 | MarkDuplicates | toolshed.g2.bx.psu.edu/repos/devteam/picard/picard_MarkDuplicates/2.18.2.2 |
| 14 | bedtools BAM to BED | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_bamtobed/2.30.0 |
| 15 | Paired-end histogram | toolshed.g2.bx.psu.edu/repos/iuc/pe_histogram/pe_histogram/1.0.1 |
| 16 | MACS2 callpeak | toolshed.g2.bx.psu.edu/repos/iuc/macs2/macs2_callpeak/2.1.1.20160309.6 |
| 17 | Wig/BedGraph-to-bigWig | wig_to_bigWig |
| 18 | computeMatrix | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_compute_matrix/deeptools_compute_matrix/3.3.2.0.0 |
| 19 | pyGenomeTracks | toolshed.g2.bx.psu.edu/repos/iuc/pygenometracks/pygenomeTracks/3.6 |
| 20 | computeMatrix | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_compute_matrix/deeptools_compute_matrix/3.3.2.0.0 |
| 21 | plotHeatmap | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_heatmap/deeptools_plot_heatmap/3.3.2.0.1 |
| 22 | plotHeatmap | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_heatmap/deeptools_plot_heatmap/3.3.2.0.1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_22 | #main/_anonymous_output_22 | n/a |
|
| _anonymous_output_23 | #main/_anonymous_output_23 | n/a |
|
| _anonymous_output_24 | #main/_anonymous_output_24 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
| computeMatrix on input dataset(s): Matrix | #main/computeMatrix on input dataset(s): Matrix | n/a |
|
| plotHeatmap on input dataset(s): Image | #main/plotHeatmap on input dataset(s): Image | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1306 Downloads: 190 Runs: 0
Created: 2nd Jun 2025 at 10:59
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master