Workflow Type: Galaxy
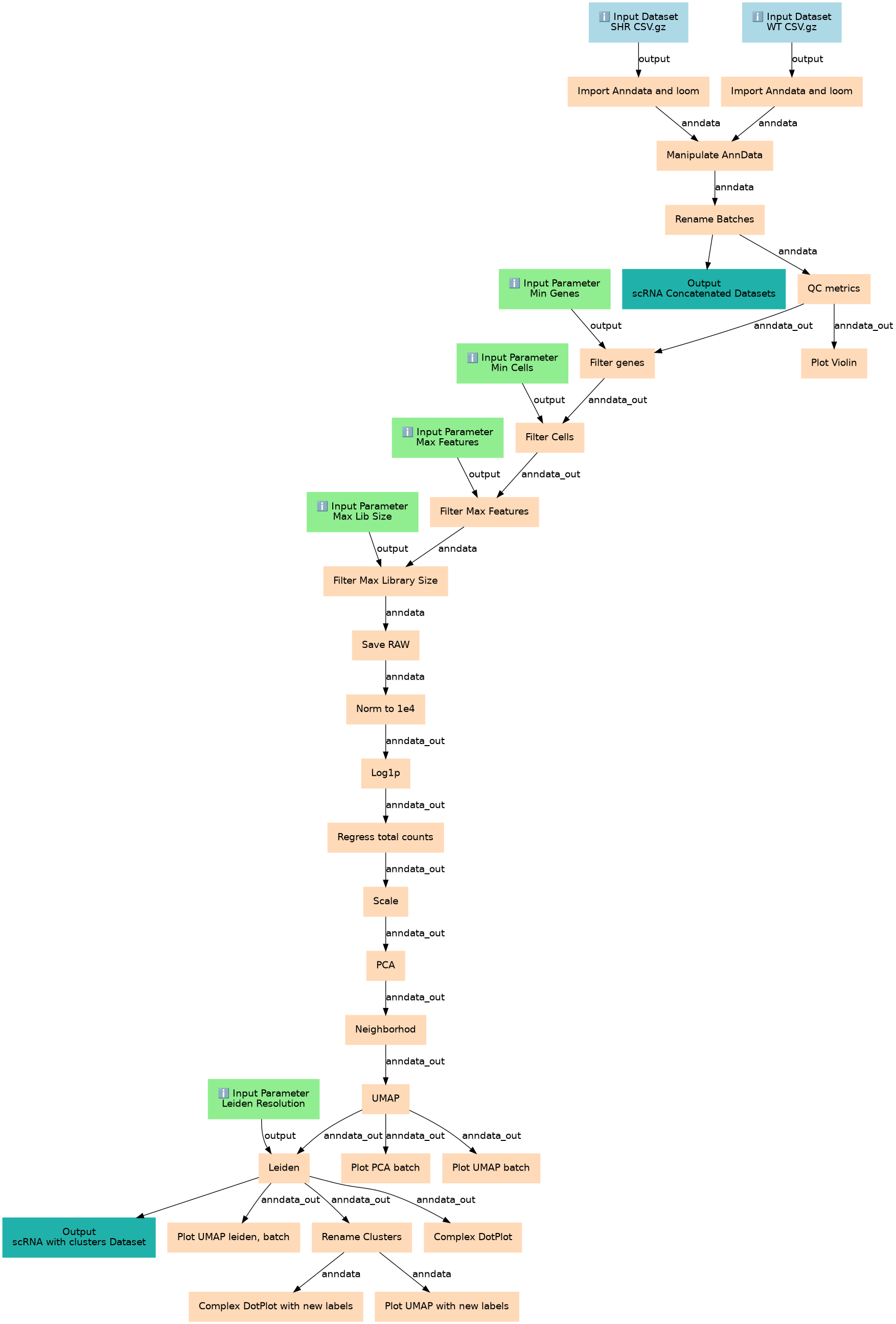
Downstream Single-cell RNA Plant analysis with ScanPy
Associated Tutorial
This workflows is part of the tutorial Analysis of plant scRNA-Seq Data with Scanpy, available in the GTN
Thanks to...
Tutorial Author(s): Mehmet Tekman, Beatriz Serrano-Solano, Cristóbal Gallardo, Pavankumar Videm
Tutorial Contributor(s): Mehmet Tekman, Wendi Bacon, Helena Rasche, Beatriz Serrano-Solano, Björn Grüning
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Leiden Resolution | #main/Leiden Resolution | n/a |
|
| Max Features | #main/Max Features | n/a |
|
| Max Lib Size | #main/Max Lib Size | n/a |
|
| Min Cells | #main/Min Cells | n/a |
|
| Min Genes | #main/Min Genes | n/a |
|
| SHR (CSV.gz) | #main/SHR (CSV.gz) | n/a |
|
| WT (CSV.gz) | #main/WT (CSV.gz) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 7 | Import Anndata and loom | toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy0 |
| 8 | Import Anndata and loom | toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy0 |
| 9 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy0 |
| 10 | Rename Batches | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy0 |
| 11 | QC metrics | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.7.1+galaxy0 |
| 12 | Filter genes | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.7.1+galaxy0 |
| 13 | Plot Violin | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
| 14 | Filter Cells | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.7.1+galaxy0 |
| 15 | Filter Max Features | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy0 |
| 16 | Filter Max Library Size | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy0 |
| 17 | Save RAW | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy0 |
| 18 | Norm to 1e4 | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_normalize/scanpy_normalize/1.7.1+galaxy0 |
| 19 | Log1p | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.7.1+galaxy0 |
| 20 | Regress total counts | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_remove_confounders/scanpy_remove_confounders/1.7.1+galaxy0 |
| 21 | Scale | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.7.1+galaxy0 |
| 22 | PCA | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.7.1+galaxy0 |
| 23 | Neighborhod | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.7.1+galaxy0 |
| 24 | UMAP | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.7.1+galaxy0 |
| 25 | Leiden | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.7.1+galaxy0 |
| 26 | Plot PCA (batch) | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
| 27 | Plot UMAP (batch) | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
| 28 | Plot UMAP (leiden, batch) | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
| 29 | Rename Clusters | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy0 |
| 30 | Complex DotPlot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
| 31 | Complex DotPlot with new labels | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
| 32 | Plot UMAP with new labels | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| scRNA Concatenated Datasets | #main/scRNA Concatenated Datasets | n/a |
|
| scRNA with clusters Dataset | #main/scRNA with clusters Dataset | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1333 Downloads: 192 Runs: 0
Created: 2nd Jun 2025 at 10:59
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master