Workflow Type: Galaxy
Associated Tutorial
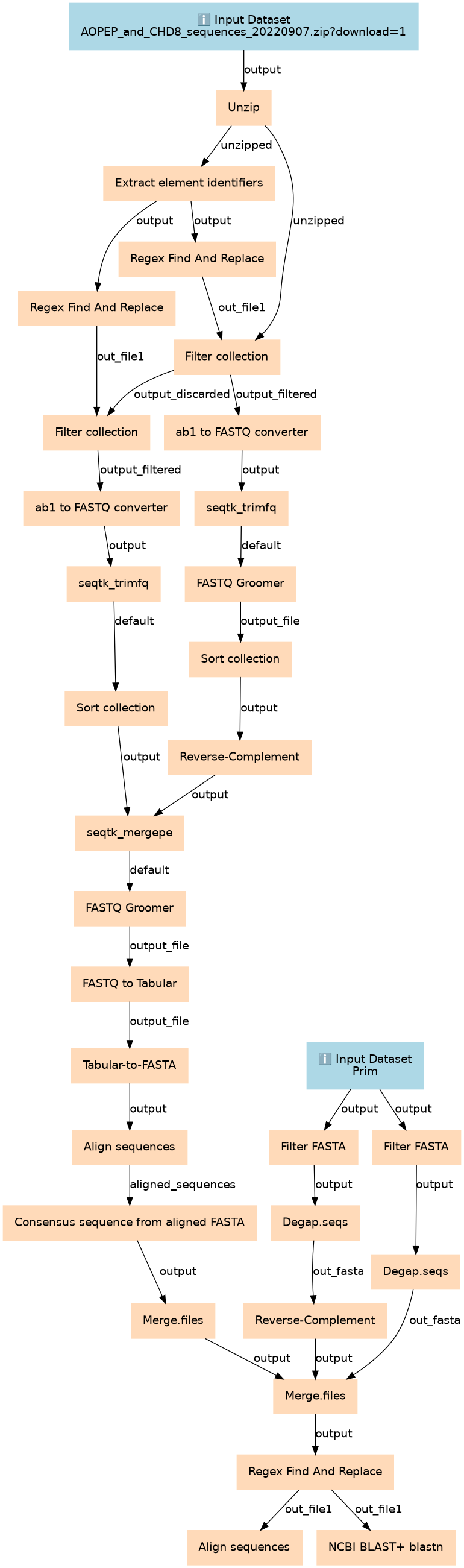
This workflows is part of the tutorial Clean and manage Sanger sequences from raw files to aligned consensus, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Coline Royaux
Tutorial Author(s): Coline Royaux
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| AOPEP_and_CHD8_sequences_20220907.zip?download=1 | AOPEP_and_CHD8_sequences_20220907.zip?download=1 | n/a |
|
| Prim | Prim | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Unzip | toolshed.g2.bx.psu.edu/repos/imgteam/unzip/unzip/6.0+galaxy0 |
| 3 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.3 |
| 4 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.3 |
| 5 | Extract element identifiers | toolshed.g2.bx.psu.edu/repos/iuc/collection_element_identifiers/collection_element_identifiers/0.0.2 |
| 6 | Degap.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_degap_seqs/mothur_degap_seqs/1.39.5.0 |
| 7 | Degap.seqs | toolshed.g2.bx.psu.edu/repos/iuc/mothur_degap_seqs/mothur_degap_seqs/1.39.5.0 |
| 8 | Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.3 |
| 9 | Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.3 |
| 10 | Reverse-Complement | toolshed.g2.bx.psu.edu/repos/devteam/fastx_reverse_complement/cshl_fastx_reverse_complement/1.0.2+galaxy0 |
| 11 | Filter collection | __FILTER_FROM_FILE__ |
| 12 | Filter collection | __FILTER_FROM_FILE__ |
| 13 | ab1 to FASTQ converter | toolshed.g2.bx.psu.edu/repos/ecology/ab1_fastq_converter/ab1_fastq_converter/1.20.0 |
| 14 | ab1 to FASTQ converter | toolshed.g2.bx.psu.edu/repos/ecology/ab1_fastq_converter/ab1_fastq_converter/1.20.0 |
| 15 | seqtk_trimfq | toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_trimfq/1.3.1 |
| 16 | seqtk_trimfq | toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_trimfq/1.3.1 |
| 17 | FASTQ Groomer | toolshed.g2.bx.psu.edu/repos/devteam/fastq_groomer/fastq_groomer/1.1.5 |
| 18 | Sort collection | __SORTLIST__ |
| 19 | Sort collection | __SORTLIST__ |
| 20 | Reverse-Complement | toolshed.g2.bx.psu.edu/repos/devteam/fastx_reverse_complement/cshl_fastx_reverse_complement/1.0.2+galaxy0 |
| 21 | seqtk_mergepe | toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_mergepe/1.3.1 |
| 22 | FASTQ Groomer | toolshed.g2.bx.psu.edu/repos/devteam/fastq_groomer/fastq_groomer/1.1.5 |
| 23 | FASTQ to Tabular | toolshed.g2.bx.psu.edu/repos/devteam/fastq_to_tabular/fastq_to_tabular/1.1.5 |
| 24 | Tabular-to-FASTA | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 25 | Align sequences | toolshed.g2.bx.psu.edu/repos/iuc/qiime_align_seqs/qiime_align_seqs/1.9.1.0 |
| 26 | Consensus sequence from aligned FASTA | toolshed.g2.bx.psu.edu/repos/ecology/aligned_to_consensus/aligned_to_consensus/1.0.0 |
| 27 | Merge.files | toolshed.g2.bx.psu.edu/repos/iuc/mothur_merge_files/mothur_merge_files/1.39.5.0 |
| 28 | Merge.files | toolshed.g2.bx.psu.edu/repos/iuc/mothur_merge_files/mothur_merge_files/1.39.5.0 |
| 29 | Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regex1/1.0.3 |
| 30 | Align sequences | toolshed.g2.bx.psu.edu/repos/iuc/qiime_align_seqs/qiime_align_seqs/1.9.1.0 |
| 31 | NCBI BLAST+ blastn | toolshed.g2.bx.psu.edu/repos/devteam/ncbi_blast_plus/ncbi_blastn_wrapper/2.10.1+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 1281 Downloads: 184 Runs: 0
Created: 2nd Jun 2025 at 11:02
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master