Workflow Type: Galaxy
Running molecular dynamics simulations using GROMACS
Associated Tutorial
This workflows is part of the tutorial Running molecular dynamics simulations using GROMACS, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Simon Bray
Steps
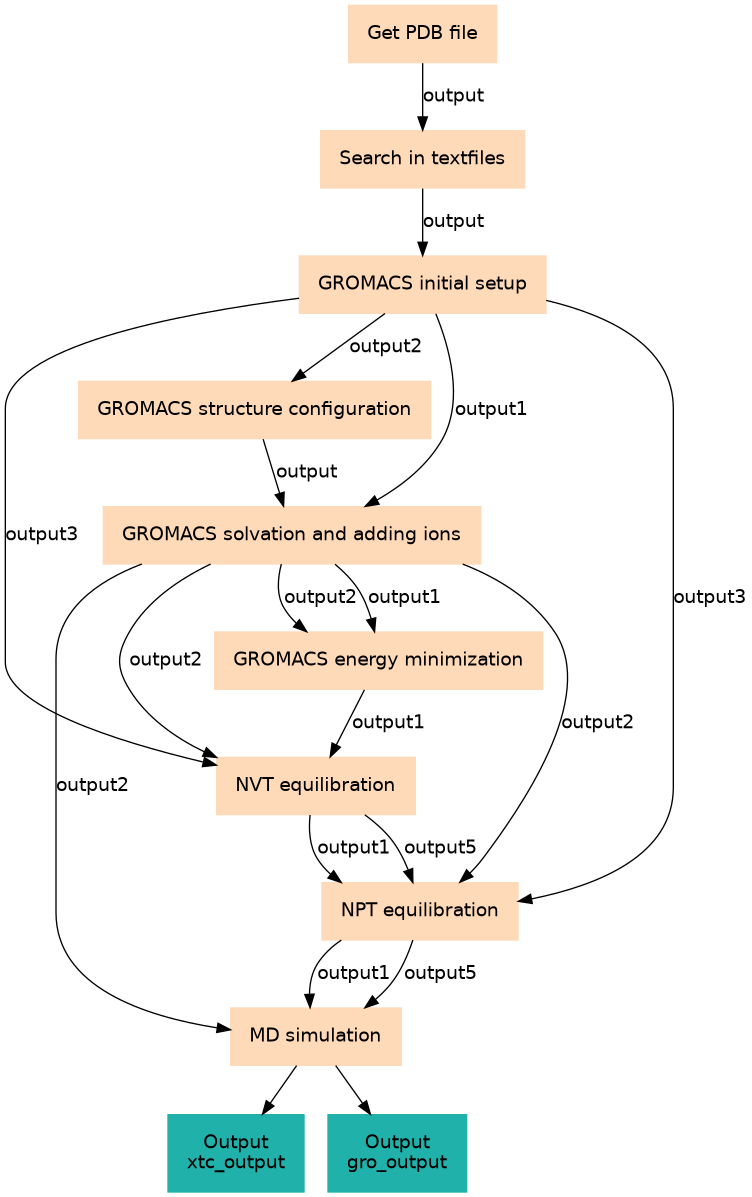
| ID | Name | Description |
|---|---|---|
| 0 | Get PDB file | toolshed.g2.bx.psu.edu/repos/bgruening/get_pdb/get_pdb/0.1.0 |
| 1 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/1.1.1 |
| 2 | GROMACS initial setup | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_setup/gmx_setup/2020.2+galaxy0 |
| 3 | GROMACS structure configuration | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_editconf/gmx_editconf/2020.2+galaxy0 |
| 4 | GROMACS solvation and adding ions | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_solvate/gmx_solvate/2020.2+galaxy0 |
| 5 | GROMACS energy minimization | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_em/gmx_em/2020.2+galaxy0 |
| 6 | NVT equilibration | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_sim/gmx_sim/2020.2+galaxy0 |
| 7 | NPT equilibration | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_sim/gmx_sim/2020.2+galaxy0 |
| 8 | MD simulation | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_sim/gmx_sim/2020.2+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| gro_output | #main/gro_output | n/a |
|
| xtc_output | #main/xtc_output | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1447 Downloads: 208 Runs: 0
Created: 2nd Jun 2025 at 11:02
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master