Workflow Type: Galaxy
Associated Tutorial
This workflows is part of the tutorial Identification and Evolutionary Analysis of Transcription-Associated Proteins in Streptophyte algae and Land plants, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Deepti Varshney, Saskia Hiltemann
Tutorial Author(s): Deepti Varshney, Saskia Hiltemann
Tutorial Contributor(s): Saskia Hiltemann, Teresa Müller
Funder(s): MAdLand - Molecular Adaptation to Land: plant evolution to change
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input Dataset Collection | Input Dataset Collection | n/a |
|
Steps
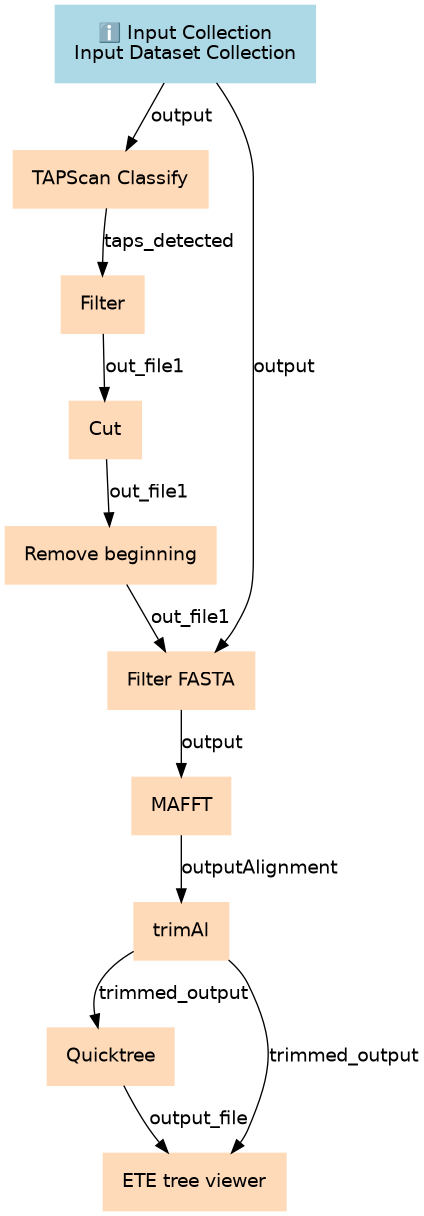
| ID | Name | Description |
|---|---|---|
| 1 | TAPScan Classify | toolshed.g2.bx.psu.edu/repos/bgruening/tapscan/tapscan_classify/4.76+galaxy0 |
| 2 | Filter | Filter1 |
| 3 | Cut | Cut1 |
| 4 | Remove beginning | Remove beginning1 |
| 5 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.3 |
| 6 | MAFFT | toolshed.g2.bx.psu.edu/repos/rnateam/mafft/rbc_mafft/7.526+galaxy1 |
| 7 | trimAl | toolshed.g2.bx.psu.edu/repos/iuc/trimal/trimal/1.5.0+galaxy1 |
| 8 | Quicktree | toolshed.g2.bx.psu.edu/repos/iuc/quicktree/quicktree/2.5+galaxy0 |
| 9 | ETE tree viewer | toolshed.g2.bx.psu.edu/repos/iuc/ete_treeviewer/ete_treeviewer/3.1.3+galaxy0 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 1303 Downloads: 198 Runs: 0
Created: 2nd Jun 2025 at 11:02
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master