Workflow Type: Galaxy
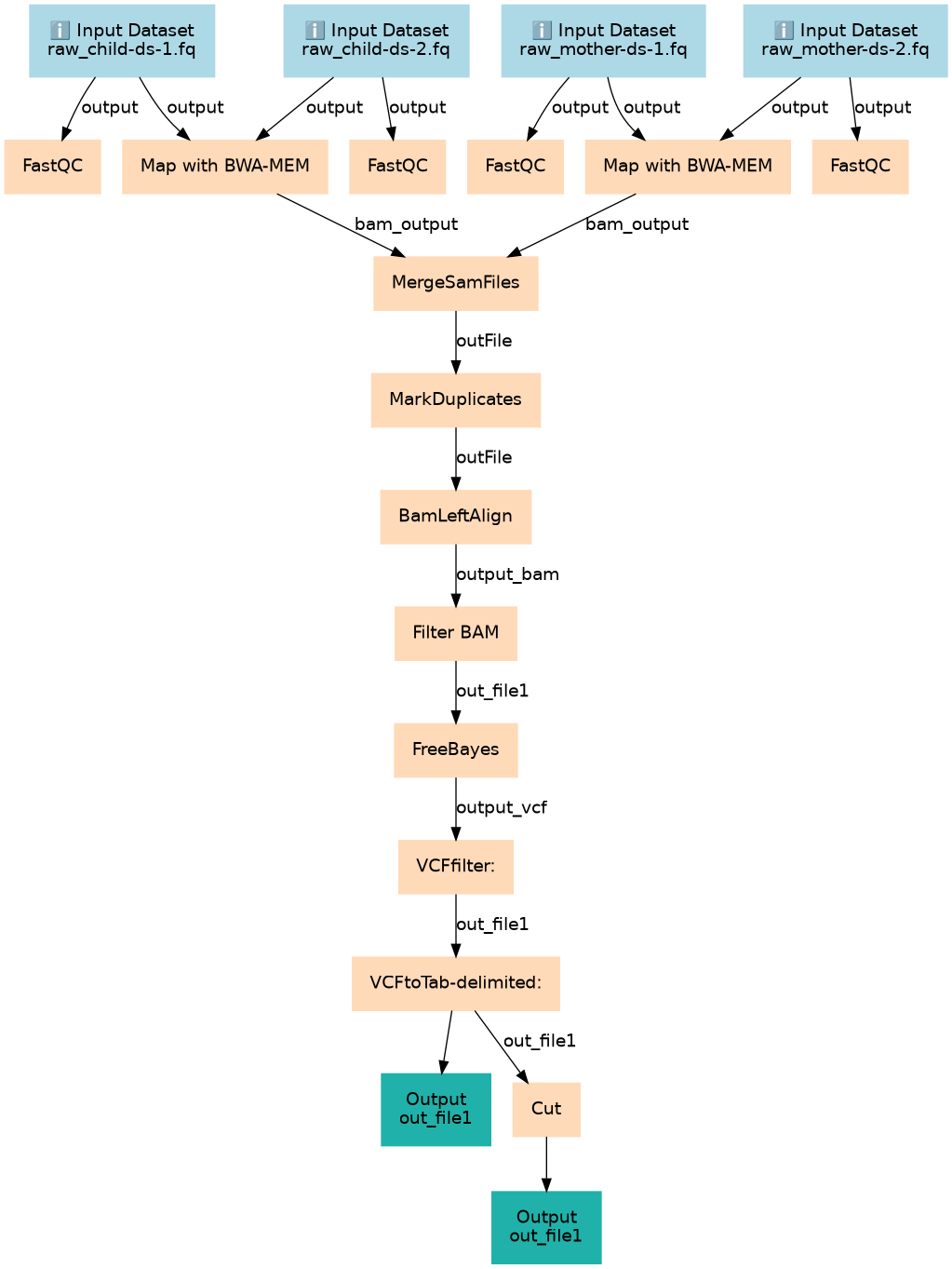
Calling variants in non-diploid systems
Associated Tutorial
This workflows is part of the tutorial Calling variants in non-diploid systems, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Anton Nekrutenko, Alex Ostrovsky
Tutorial Author(s): Anton Nekrutenko, Alex Ostrovsky
Steps
| ID | Name | Description |
|---|---|---|
| 4 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 5 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 6 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.19 |
| 7 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 8 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 9 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.19 |
| 10 | MergeSamFiles | toolshed.g2.bx.psu.edu/repos/devteam/picard/picard_MergeSamFiles/3.1.1.0 |
| 11 | MarkDuplicates | toolshed.g2.bx.psu.edu/repos/devteam/picard/picard_MarkDuplicates/3.1.1.0 |
| 12 | BamLeftAlign | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/bamleftalign/1.3.9 |
| 13 | Filter BAM | toolshed.g2.bx.psu.edu/repos/devteam/bamtools_filter/bamFilter/2.5.2+galaxy2 |
| 14 | FreeBayes | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/freebayes/1.3.9+galaxy0 |
| 15 | VCFfilter: | toolshed.g2.bx.psu.edu/repos/devteam/vcffilter/vcffilter2/1.0.0_rc3+galaxy3 |
| 16 | VCFtoTab-delimited: | toolshed.g2.bx.psu.edu/repos/devteam/vcf2tsv/vcf2tsv/1.0.0_rc3+galaxy0 |
| 17 | Cut | Cut1 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1332 Downloads: 187 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master