Workflow Type: Galaxy
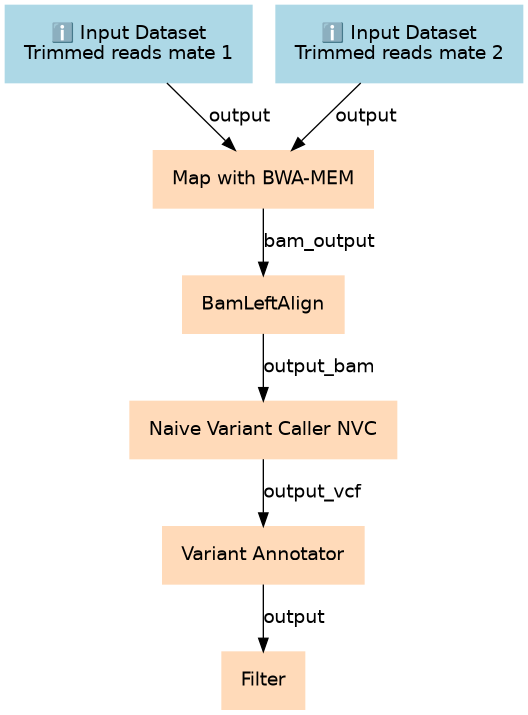
Run this on the trimmed consensus reads (DCS or SSCS) from Du Novo.
Associated Tutorial
This workflows is part of the tutorial Calling very rare variants, available in the GTN
Thanks to...
Tutorial Author(s): Anton Nekrutenko, Nick Stoler
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Trimmed reads (mate 1) | #main/Trimmed reads (mate 1) | n/a |
|
| Trimmed reads (mate 2) | #main/Trimmed reads (mate 2) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.17.1 |
| 3 | BamLeftAlign | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/bamleftalign/1.1.0.46-0 |
| 4 | Naive Variant Caller (NVC) | toolshed.g2.bx.psu.edu/repos/blankenberg/naive_variant_caller/naive_variant_caller/0.0.4 |
| 5 | Variant Annotator | toolshed.g2.bx.psu.edu/repos/nick/allele_counts/allele_counts_1/1.2 |
| 6 | Filter | Filter1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1140 Downloads: 167 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master