Workflow Type: Galaxy
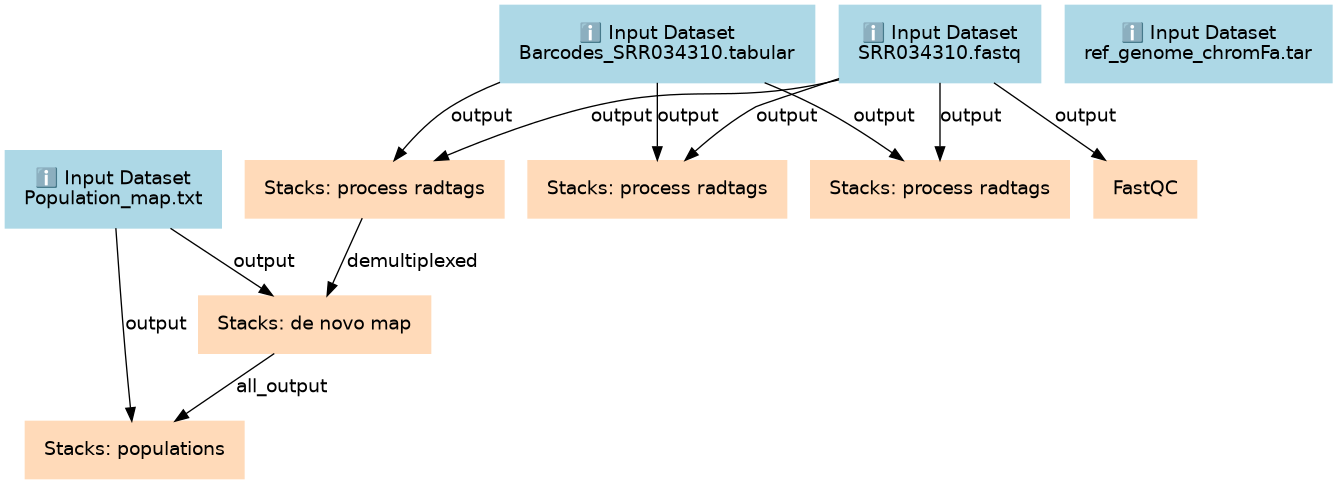
RAD-Seq de-novo data analysis
Associated Tutorial
This workflows is part of the tutorial RAD-Seq de-novo data analysis, available in the GTN
Thanks to...
Tutorial Author(s): Yvan Le Bras
Tutorial Contributor(s): Verena Moosmann, Helena Rasche, Saskia Hiltemann, Nicola Soranzo, Björn Grüning, Yvan Le Bras, Gildas Le Corguillé, Bérénice Batut, William Durand, Niall Beard, Wolfgang Maier
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Barcodes_SRR034310.tabular | Barcodes_SRR034310.tabular | n/a |
|
| Population_map.txt | Population_map.txt | n/a |
|
| SRR034310.fastq | SRR034310.fastq | n/a |
|
| ref_genome_chromFa.tar | ref_genome_chromFa.tar | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Stacks: process radtags | toolshed.g2.bx.psu.edu/repos/iuc/stacks_procrad/stacks_procrad/1.46.0 |
| 5 | Stacks: process radtags | toolshed.g2.bx.psu.edu/repos/iuc/stacks_procrad/stacks_procrad/1.46.0 |
| 6 | Stacks: process radtags | toolshed.g2.bx.psu.edu/repos/iuc/stacks_procrad/stacks_procrad/1.46.0 |
| 7 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.69 |
| 8 | Stacks: de novo map | toolshed.g2.bx.psu.edu/repos/iuc/stacks_denovomap/stacks_denovomap/1.46.0 |
| 9 | Stacks: populations | toolshed.g2.bx.psu.edu/repos/iuc/stacks_populations/stacks_populations/1.46.0 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1319 Downloads: 181 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master