Workflow Type: Galaxy
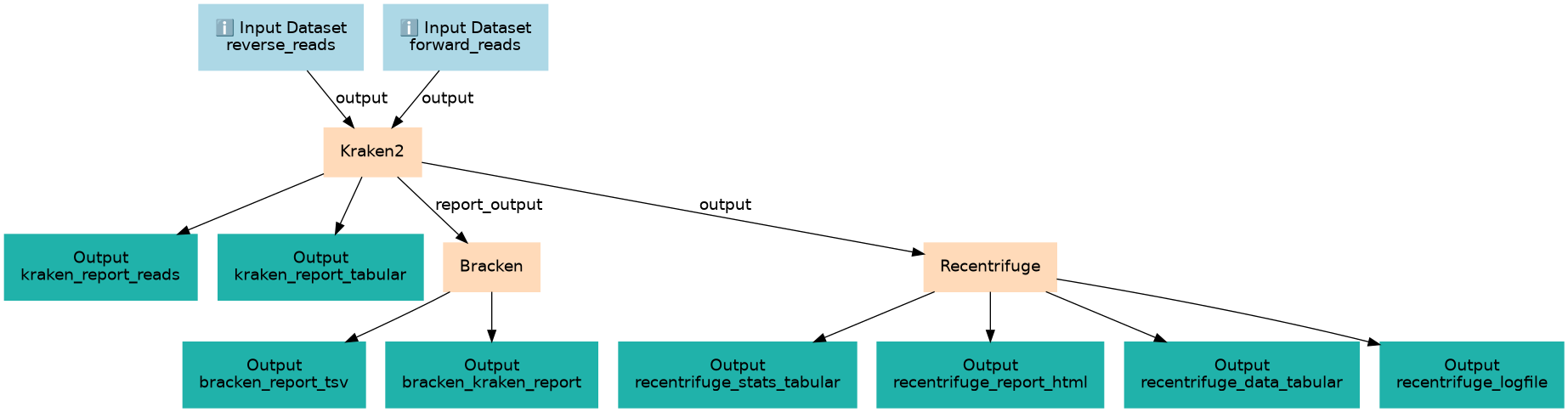
Checking expected species and contamination in bacterial isolate
Associated Tutorial
This workflows is part of the tutorial Checking expected species and contamination in bacterial isolate, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Pierre, Bérénice Batut
Tutorial Author(s): Bérénice Batut
Tutorial Contributor(s): Clea Siguret, Helena Rasche, Saskia Hiltemann
Funder(s): ABRomics
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| forward_reads | #main/forward_reads | forward_reads |
|
| reverse_reads | #main/reverse_reads | reverse_reads |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Kraken2 | toolshed.g2.bx.psu.edu/repos/iuc/kraken2/kraken2/2.1.1+galaxy1 |
| 3 | Bracken | toolshed.g2.bx.psu.edu/repos/iuc/bracken/est_abundance/2.9+galaxy0 |
| 4 | Recentrifuge | toolshed.g2.bx.psu.edu/repos/iuc/recentrifuge/recentrifuge/1.12.1+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| bracken_kraken_report | #main/bracken_kraken_report | n/a |
|
| bracken_report_tsv | #main/bracken_report_tsv | n/a |
|
| kraken_report_reads | #main/kraken_report_reads | n/a |
|
| kraken_report_tabular | #main/kraken_report_tabular | n/a |
|
| recentrifuge_data_tabular | #main/recentrifuge_data_tabular | n/a |
|
| recentrifuge_logfile | #main/recentrifuge_logfile | n/a |
|
| recentrifuge_report_html | #main/recentrifuge_report_html | n/a |
|
| recentrifuge_stats_tabular | #main/recentrifuge_stats_tabular | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 1276 Downloads: 211 Runs: 0
Created: 2nd Jun 2025 at 11:03
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master