Workflow Type: Galaxy
Reproduce the tutorial analysis at https://training.galaxyproject.org/training-material/topics/variant-analysis/tutorials/mapping-by-sequencing/tutorial.html
Associated Tutorial
This workflows is part of the tutorial Mapping and molecular identification of phenotype-causing mutations, available in the GTN
Thanks to...
Tutorial Author(s): Wolfgang Maier
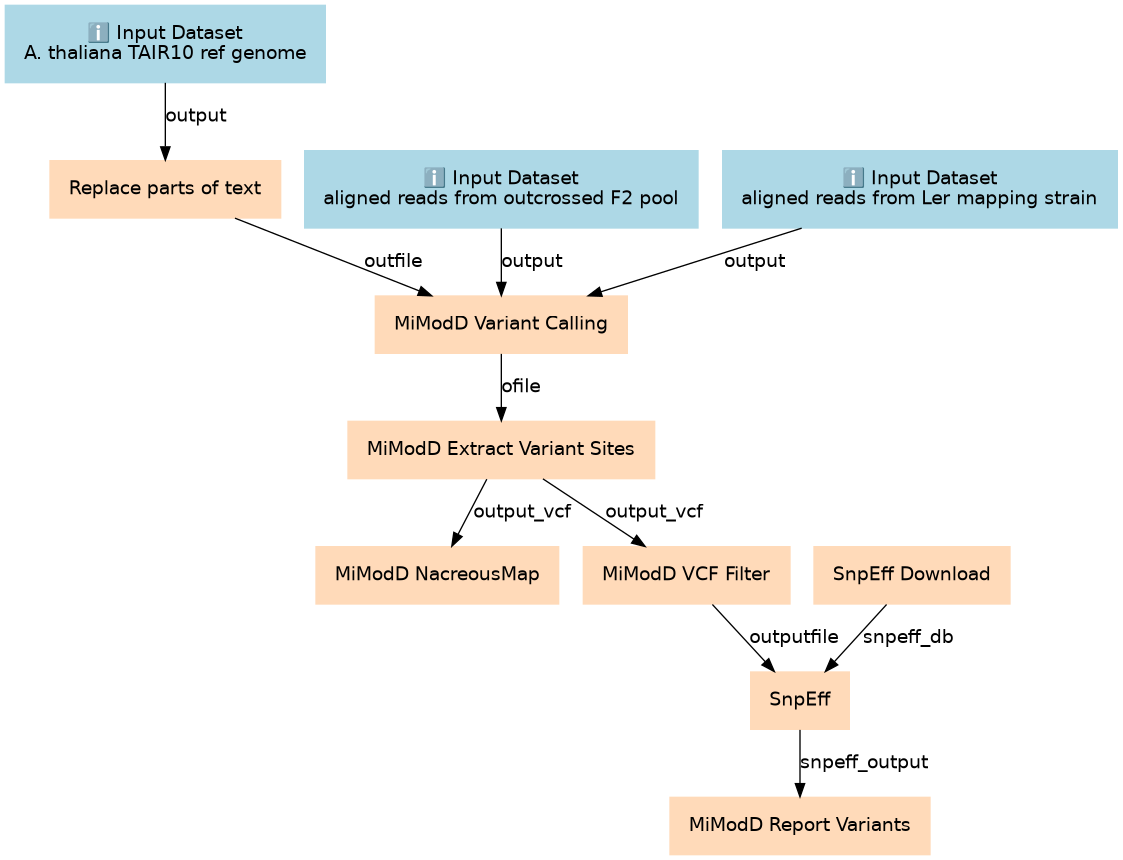
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| A. thaliana TAIR10 ref genome | A. thaliana TAIR10 ref genome | can be obtained from https://www.arabidopsis.org/download_files/Genes/TAIR10_genome_release/TAIR10_chromosome_files/TAIR10_chr_all.fas |
|
| aligned reads from Ler mapping strain | aligned reads from Ler mapping strain | as obtained from https://zenodo.org/record/1098034/files/Ler_mapping_strain.bam |
|
| aligned reads from outcrossed F2 pool | aligned reads from outcrossed F2 pool | as obtained from https://zenodo.org/record/1098034/files/outcrossed_F2.bam |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | SnpEff Download | download the SnpEff genome annotation file corresponding to TAIR10 toolshed.g2.bx.psu.edu/repos/iuc/snpeff/snpEff_download/4.1.0 |
| 4 | Replace parts of text | truncate ref genome chromosome names toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/1.1.1 |
| 5 | MiModD Variant Calling | compute variant call statistics for both samples across the entire genome toolshed.g2.bx.psu.edu/repos/wolma/mimodd_main/mimodd_varcall/0.1.8_1 |
| 6 | MiModD Extract Variant Sites | extract variants at sites for which at least one sample is not homozygous wildtype toolshed.g2.bx.psu.edu/repos/wolma/mimodd_main/mimodd_varextract/0.1.8_1 |
| 7 | MiModD NacreousMap | Variant allele frequency-based linkage analysis toolshed.g2.bx.psu.edu/repos/wolma/mimodd_main/mimodd_map/0.1.8_1 |
| 8 | MiModD VCF Filter | get candidates in linked region toolshed.g2.bx.psu.edu/repos/wolma/mimodd_main/mimodd_vcf_filter/0.1.8_1 |
| 9 | SnpEff | annotate the candidate mutations toolshed.g2.bx.psu.edu/repos/iuc/snpeff/snpEff/4.1.0 |
| 10 | MiModD Report Variants | toolshed.g2.bx.psu.edu/repos/wolma/mimodd_main/mimodd_varreport/0.1.8_1 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1386 Downloads: 203 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master