Workflow Type: Galaxy
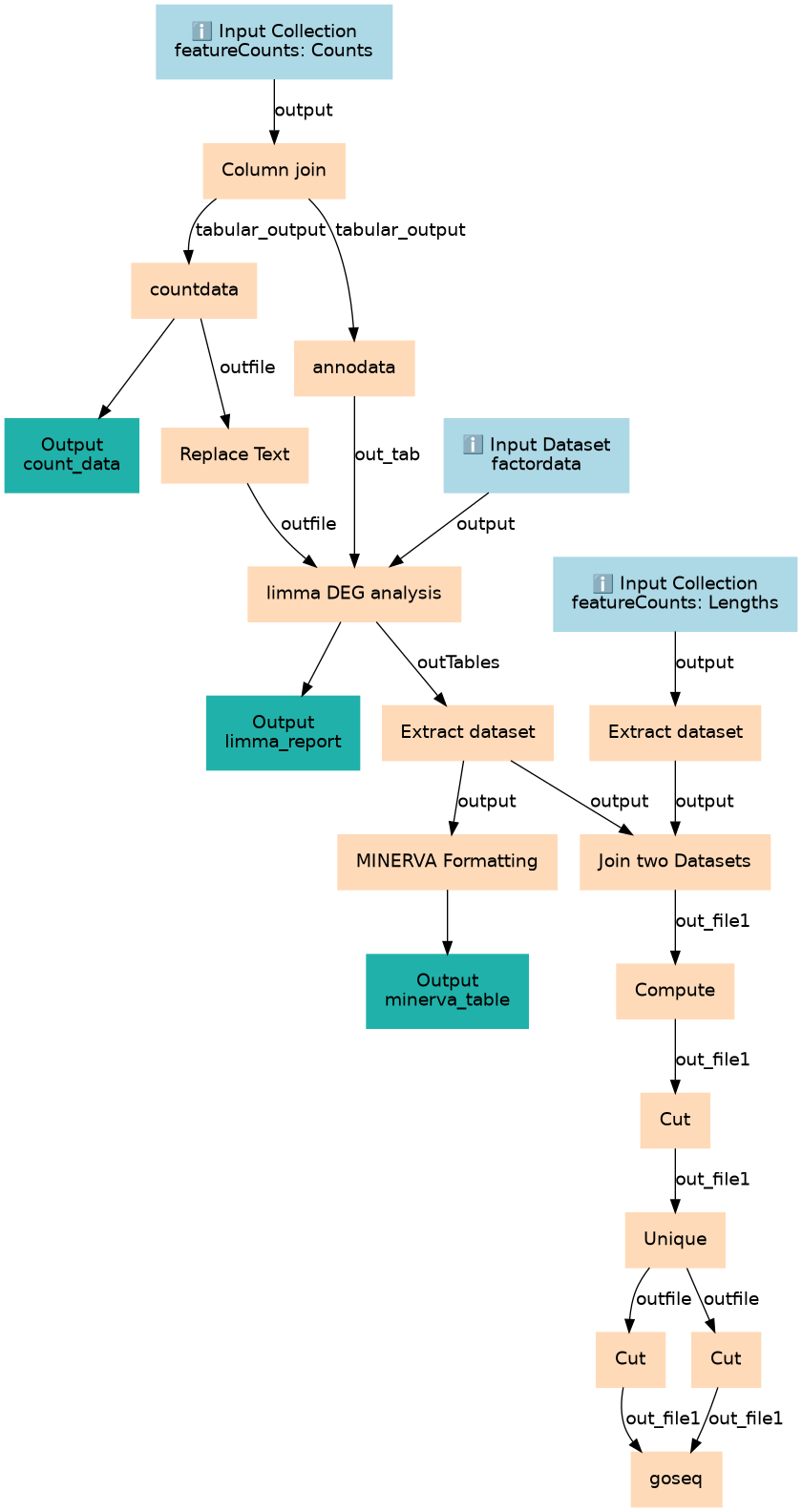
Analyse Bulk RNA-Seq data in preparation for downstream Pathways analysis with MINERVA
Associated Tutorial
This workflows is part of the tutorial Pathway analysis with the MINERVA Platform, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Iacopo Cristoferi, Helena Rasche, Clinical Bioinformatics Unit, Pathology Department, Eramus Medical Center
Tutorial Author(s): Marek Ostaszewski, Matti Hoch, Iacopo Cristoferi, Myrthe van Baardwijk, Saskia Hiltemann, Helena Rasche
Tutorial Contributor(s): Helena Rasche, Matti Hoch, Mira Kuntz, José Manuel Domínguez, Sanjay Kumar Srikakulam, Björn Grüning, Linelle Abueg, Saskia Hiltemann
Grants(s): BeYond-COVID
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| factordata | factordata | A two column factor table with (Sample Identifier, Condition) This workflow assumes a 1 factor, 2 level analysis, and was specifically designed around SARS-CoV-2 analysis with two levels, e.g. ``` SampleName Group SRR16683284 COVID SRR16683283 COVID SRR16683271 healthy SRR16683270 healthy ``` |
|
| featureCounts: Counts | featureCounts: Counts | count data collection with two column datasets (gene_id, count) |
|
| featureCounts: Lengths | featureCounts: Lengths | featureCounts Lengths collection |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Column join | toolshed.g2.bx.psu.edu/repos/iuc/collection_column_join/collection_column_join/0.0.3 |
| 4 | Extract dataset | __EXTRACT_DATASET__ |
| 5 | countdata | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/1.1.1 |
| 6 | annodata | toolshed.g2.bx.psu.edu/repos/iuc/annotatemyids/annotatemyids/3.16.0+galaxy1 |
| 7 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_line/1.1.2 |
| 8 | limma DEG analysis | toolshed.g2.bx.psu.edu/repos/iuc/limma_voom/limma_voom/3.50.1+galaxy0 |
| 9 | Extract dataset | __EXTRACT_DATASET__ |
| 10 | MINERVA Formatting | Cut1 |
| 11 | Join two Datasets | join1 |
| 12 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/1.4 |
| 13 | Cut | Cut1 |
| 14 | Unique | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sorted_uniq/1.1.0 |
| 15 | Cut | Cut1 |
| 16 | Cut | Cut1 |
| 17 | goseq | toolshed.g2.bx.psu.edu/repos/iuc/goseq/goseq/1.50.0+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| count_data | count_data | n/a |
|
| limma_report | limma_report | n/a |
|
| minerva_table | minerva_table | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 1318 Downloads: 190 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master