Workflow Type: Galaxy
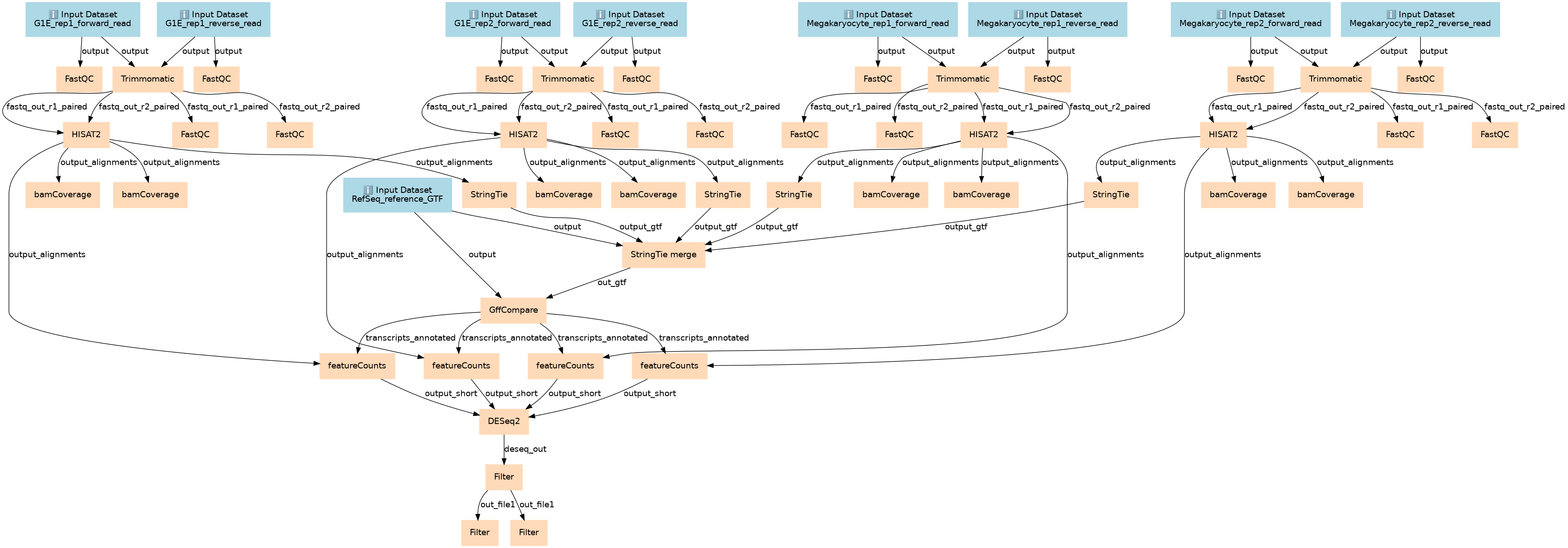
De novo transcriptome reconstruction with RNA-Seq
Associated Tutorial
This workflows is part of the tutorial De novo transcriptome reconstruction with RNA-Seq, available in the GTN
Thanks to...
Tutorial Author(s): Mallory Freeberg, Mo Heydarian
Tutorial Contributor(s): James Taylor, Saskia Hiltemann, Nicola Soranzo, Helena Rasche, Anthony Bretaudeau, Gildas Le Corguillé, Björn Grüning, Mallory Freeberg, Mo Heydarian, Bérénice Batut, William Durand, Niall Beard
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| G1E_rep1_forward_read | #main/G1E_rep1_forward_read | n/a |
|
| G1E_rep1_reverse_read | #main/G1E_rep1_reverse_read | n/a |
|
| G1E_rep2_forward_read | #main/G1E_rep2_forward_read | n/a |
|
| G1E_rep2_reverse_read | #main/G1E_rep2_reverse_read | n/a |
|
| Megakaryocyte_rep1_forward_read | #main/Megakaryocyte_rep1_forward_read | n/a |
|
| Megakaryocyte_rep1_reverse_read | #main/Megakaryocyte_rep1_reverse_read | n/a |
|
| Megakaryocyte_rep2_forward_read | #main/Megakaryocyte_rep2_forward_read | n/a |
|
| Megakaryocyte_rep2_reverse_read | #main/Megakaryocyte_rep2_reverse_read | n/a |
|
| RefSeq_reference_GTF | #main/RefSeq_reference_GTF | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 9 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 10 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 11 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 12 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 13 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 14 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 15 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 16 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 17 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 18 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 19 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 20 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 21 | HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.1.0+galaxy5 |
| 22 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 23 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 24 | HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.1.0+galaxy5 |
| 25 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 26 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 27 | HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.1.0+galaxy5 |
| 28 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 29 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 30 | HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.1.0+galaxy5 |
| 31 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 32 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 33 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/1.3.6 |
| 34 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 35 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 36 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/1.3.6 |
| 37 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 38 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 39 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/1.3.6 |
| 40 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 41 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 42 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/1.3.6 |
| 43 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 44 | bamCoverage | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_coverage/deeptools_bam_coverage/3.3.0.0.0 |
| 45 | StringTie merge | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie_merge/1.3.6 |
| 46 | GffCompare | toolshed.g2.bx.psu.edu/repos/iuc/gffcompare/gffcompare/0.11.2 |
| 47 | featureCounts | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/1.6.4+galaxy1 |
| 48 | featureCounts | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/1.6.4+galaxy1 |
| 49 | featureCounts | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/1.6.4+galaxy1 |
| 50 | featureCounts | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/1.6.4+galaxy1 |
| 51 | DESeq2 | toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.40.6 |
| 52 | Filter | Filter1 |
| 53 | Filter | Filter1 |
| 54 | Filter | Filter1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_22 | #main/_anonymous_output_22 | n/a |
|
| _anonymous_output_23 | #main/_anonymous_output_23 | n/a |
|
| _anonymous_output_24 | #main/_anonymous_output_24 | n/a |
|
| _anonymous_output_25 | #main/_anonymous_output_25 | n/a |
|
| _anonymous_output_26 | #main/_anonymous_output_26 | n/a |
|
| _anonymous_output_27 | #main/_anonymous_output_27 | n/a |
|
| _anonymous_output_28 | #main/_anonymous_output_28 | n/a |
|
| _anonymous_output_29 | #main/_anonymous_output_29 | n/a |
|
| _anonymous_output_30 | #main/_anonymous_output_30 | n/a |
|
| _anonymous_output_31 | #main/_anonymous_output_31 | n/a |
|
| _anonymous_output_32 | #main/_anonymous_output_32 | n/a |
|
| _anonymous_output_33 | #main/_anonymous_output_33 | n/a |
|
| _anonymous_output_34 | #main/_anonymous_output_34 | n/a |
|
| _anonymous_output_35 | #main/_anonymous_output_35 | n/a |
|
| _anonymous_output_36 | #main/_anonymous_output_36 | n/a |
|
| _anonymous_output_37 | #main/_anonymous_output_37 | n/a |
|
| _anonymous_output_38 | #main/_anonymous_output_38 | n/a |
|
| _anonymous_output_39 | #main/_anonymous_output_39 | n/a |
|
| _anonymous_output_40 | #main/_anonymous_output_40 | n/a |
|
| _anonymous_output_41 | #main/_anonymous_output_41 | n/a |
|
| _anonymous_output_42 | #main/_anonymous_output_42 | n/a |
|
| _anonymous_output_43 | #main/_anonymous_output_43 | n/a |
|
| _anonymous_output_44 | #main/_anonymous_output_44 | n/a |
|
| _anonymous_output_45 | #main/_anonymous_output_45 | n/a |
|
| _anonymous_output_46 | #main/_anonymous_output_46 | n/a |
|
| _anonymous_output_47 | #main/_anonymous_output_47 | n/a |
|
| _anonymous_output_48 | #main/_anonymous_output_48 | n/a |
|
| _anonymous_output_49 | #main/_anonymous_output_49 | n/a |
|
| _anonymous_output_50 | #main/_anonymous_output_50 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1327 Downloads: 186 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master