Workflow Type: Galaxy
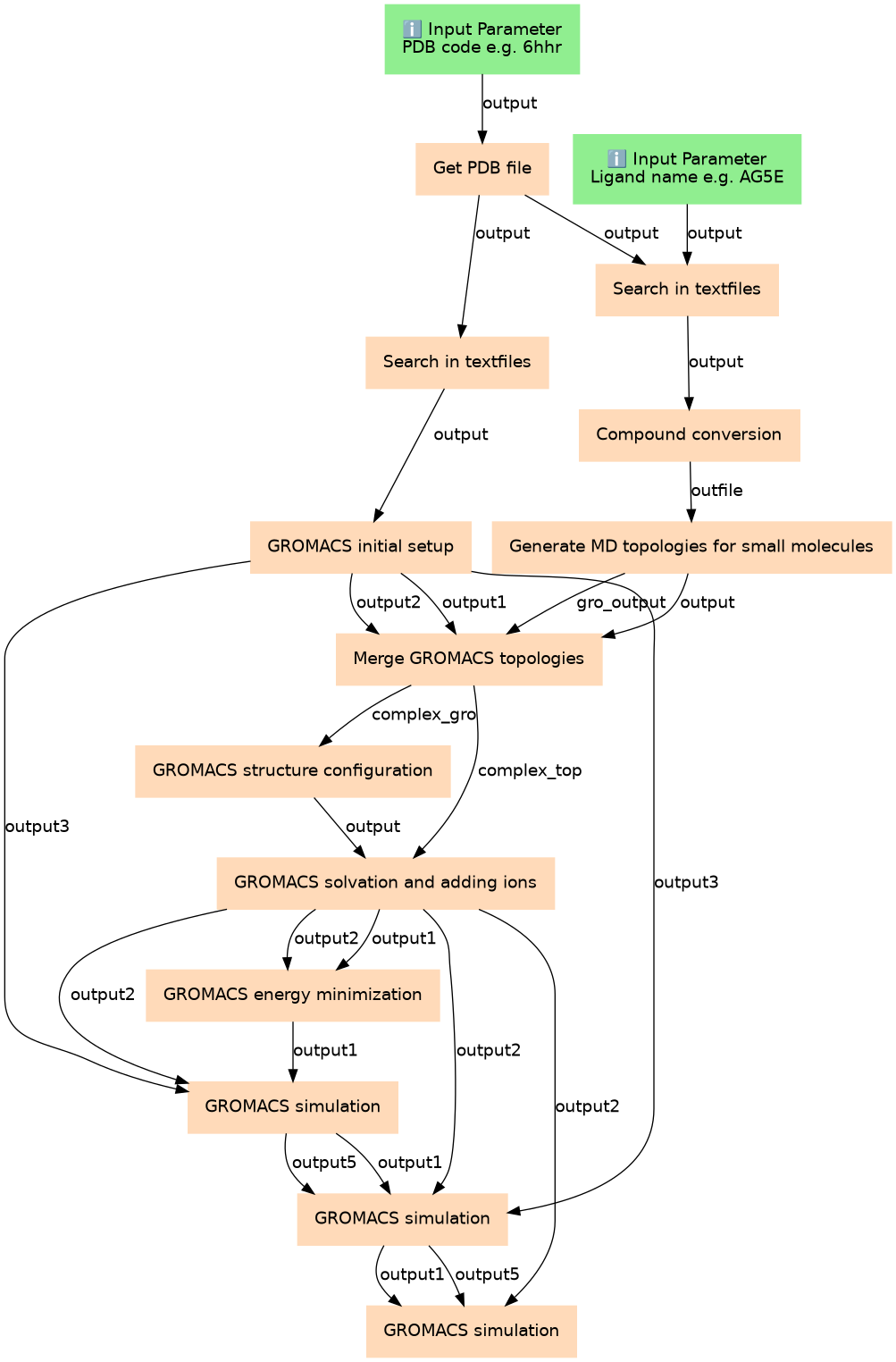
High Throughput Molecular Dynamics and Analysis
Associated Tutorial
This workflows is part of the tutorial High Throughput Molecular Dynamics and Analysis, available in the GTN
Thanks to...
Tutorial Author(s): Simon Bray, Tharindu Senapathi, Christopher Barnett, Björn Grüning
Tutorial Contributor(s): Helena Rasche, Nadia Goué, Saskia Hiltemann, Simon Bray, Simon Gladman, Christopher Barnett, Tharindu Senapathi, Stéphanie Robin, Anthony Bretaudeau, Martin Čech, Björn Grüning, Armin Dadras
Funder(s): ELIXIR Europe, de.NBI, University of Freiburg
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Ligand name (e.g. AG5E) | Ligand name (e.g. AG5E) | n/a |
|
| PDB code (e.g. 6hhr) | PDB code (e.g. 6hhr) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Get PDB file | toolshed.g2.bx.psu.edu/repos/bgruening/get_pdb/get_pdb/0.1.0 |
| 3 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/1.1.1 |
| 4 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/1.1.1 |
| 5 | GROMACS initial setup | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_setup/gmx_setup/2019.1.4 |
| 6 | Compound conversion | toolshed.g2.bx.psu.edu/repos/bgruening/openbabel_compound_convert/openbabel_compound_convert/2.4.2.2.0 |
| 7 | Generate MD topologies for small molecules | toolshed.g2.bx.psu.edu/repos/chemteam/ambertools_acpype/ambertools_acpype/19.11 |
| 8 | Merge GROMACS topologies | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_merge_topology_files/gmx_merge_topology_files/3.2.0 |
| 9 | GROMACS structure configuration | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_editconf/gmx_editconf/2019.1.4 |
| 10 | GROMACS solvation and adding ions | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_solvate/gmx_solvate/2019.1.4 |
| 11 | GROMACS energy minimization | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_em/gmx_em/2019.1.4 |
| 12 | GROMACS simulation | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_sim/gmx_sim/2019.1.4.1 |
| 13 | GROMACS simulation | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_sim/gmx_sim/2019.1.4.1 |
| 14 | GROMACS simulation | toolshed.g2.bx.psu.edu/repos/chemteam/gmx_sim/gmx_sim/2019.1.4.1 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1423 Downloads: 186 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master