Workflow Type: Galaxy
rna-seq-reads-to-counts
Associated Tutorial
This workflows is part of the tutorial 1: RNA-Seq reads to counts, available in the GTN
Thanks to...
Tutorial Author(s): Maria Doyle, Belinda Phipson, Harriet Dashnow
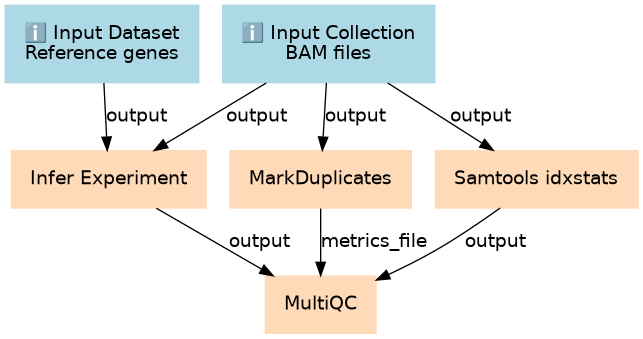
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| BAM files | #main/BAM files | n/a |
|
| Reference genes | #main/Reference genes | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Infer Experiment | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_infer_experiment/2.6.4.1 |
| 3 | MarkDuplicates | toolshed.g2.bx.psu.edu/repos/devteam/picard/picard_MarkDuplicates/2.18.2.1 |
| 4 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.2 |
| 5 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.7 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1266 Downloads: 219 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master