Workflow Type: Galaxy
roscoff hackathon
Associated Tutorial
This workflows is part of the tutorial De novo transcriptome assembly, annotation, and differential expression analysis, available in the GTN
Thanks to...
Tutorial Author(s): Anthony Bretaudeau, Gildas Le Corguillé, Erwan Corre, Xi Liu
Inputs
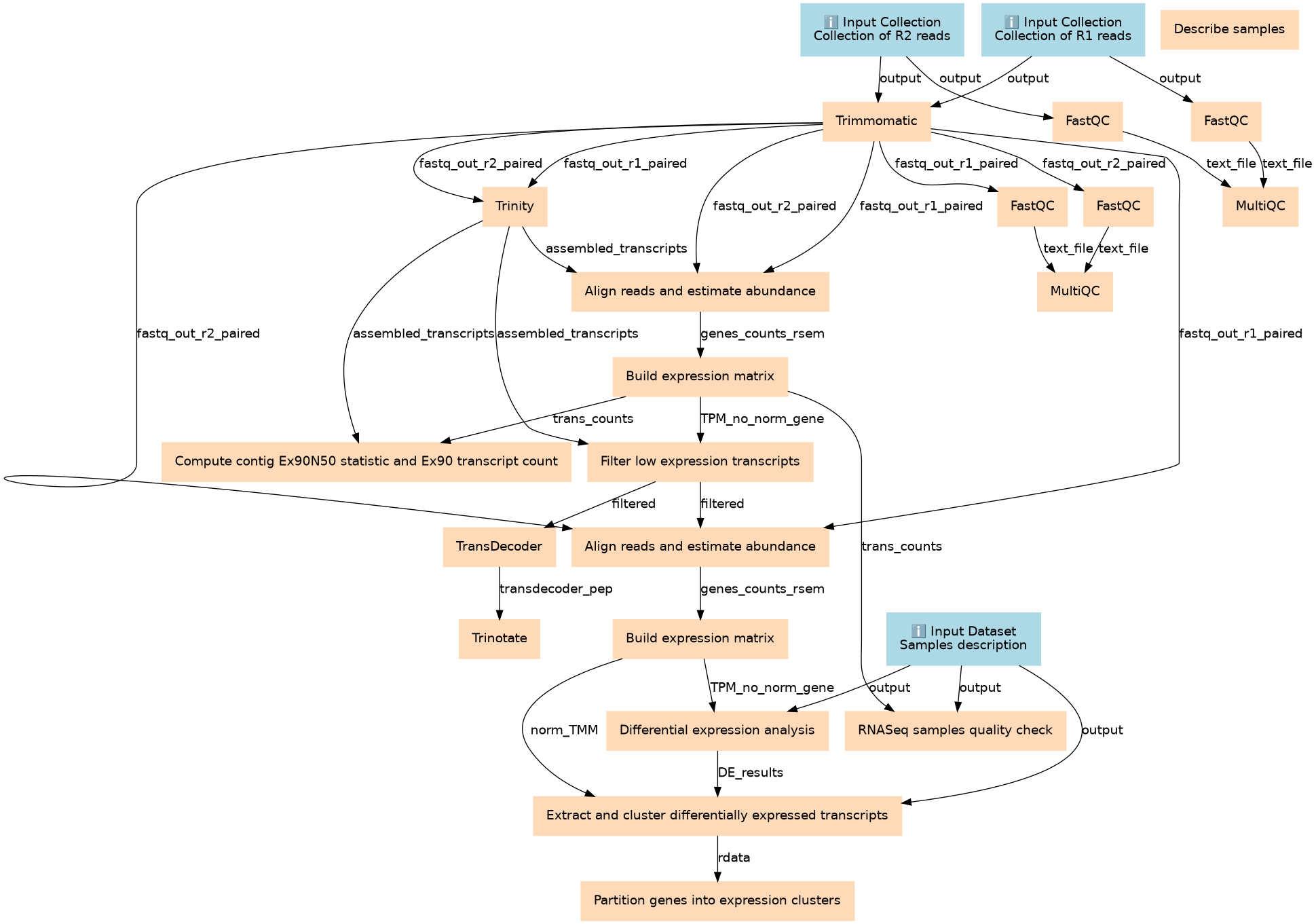
| ID | Name | Description | Type |
|---|---|---|---|
| Collection of R1 reads | Collection of R1 reads | n/a |
|
| Collection of R2 reads | Collection of R2 reads | n/a |
|
| Samples description | Samples description | The list of samples and replicates. Can be created with the 'Describe samples and replicates' tool |
|
Steps
| ID | Name | Description |
|---|---|---|
| 20 | Trinotate | toolshed.g2.bx.psu.edu/repos/iuc/trinotate/trinotate/3.1.1 |
| 21 | Differential expression analysis | toolshed.g2.bx.psu.edu/repos/iuc/trinity_run_de_analysis/trinity_run_de_analysis/2.8.4 |
| 22 | Extract and cluster differentially expressed transcripts | toolshed.g2.bx.psu.edu/repos/iuc/trinity_analyze_diff_expr/trinity_analyze_diff_expr/2.8.4 |
| 23 | Partition genes into expression clusters | toolshed.g2.bx.psu.edu/repos/iuc/trinity_define_clusters_by_cutting_tree/trinity_define_clusters_by_cutting_tree/2.8.4 |
| 2 | Describe samples | toolshed.g2.bx.psu.edu/repos/iuc/describe_samples/describe_samples/2.8.4 |
| 5 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.69 |
| 4 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.69 |
| 7 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.2.0 |
| 6 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.32.3 |
| 9 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.69 |
| 8 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.69 |
| 11 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.2.0 |
| 10 | Trinity | toolshed.g2.bx.psu.edu/repos/iuc/trinity/trinity/2.8.4 |
| 13 | Build expression matrix | toolshed.g2.bx.psu.edu/repos/iuc/trinity_abundance_estimates_to_matrix/trinity_abundance_estimates_to_matrix/2.8.4 |
| 12 | Align reads and estimate abundance | toolshed.g2.bx.psu.edu/repos/iuc/trinity_align_and_estimate_abundance/trinity_align_and_estimate_abundance/2.8.4 |
| 15 | Filter low expression transcripts | toolshed.g2.bx.psu.edu/repos/iuc/trinity_filter_low_expr_transcripts/trinity_filter_low_expr_transcripts/2.8.4 |
| 14 | RNASeq samples quality check | toolshed.g2.bx.psu.edu/repos/iuc/trinity_samples_qccheck/trinity_samples_qccheck/2.8.4 |
| 17 | Align reads and estimate abundance | toolshed.g2.bx.psu.edu/repos/iuc/trinity_align_and_estimate_abundance/trinity_align_and_estimate_abundance/2.8.4 |
| 16 | Compute contig Ex90N50 statistic and Ex90 transcript count | toolshed.g2.bx.psu.edu/repos/iuc/trinity_contig_exn50_statistic/trinity_contig_exn50_statistic/2.8.4 |
| 19 | Build expression matrix | toolshed.g2.bx.psu.edu/repos/iuc/trinity_abundance_estimates_to_matrix/trinity_abundance_estimates_to_matrix/2.8.4 |
| 18 | TransDecoder | toolshed.g2.bx.psu.edu/repos/iuc/transdecoder/transdecoder/3.0.1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| _anonymous_output_11 | _anonymous_output_11 | n/a |
|
| _anonymous_output_12 | _anonymous_output_12 | n/a |
|
| _anonymous_output_13 | _anonymous_output_13 | n/a |
|
| _anonymous_output_14 | _anonymous_output_14 | n/a |
|
| _anonymous_output_15 | _anonymous_output_15 | n/a |
|
| _anonymous_output_16 | _anonymous_output_16 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1402 Downloads: 195 Runs: 0
Created: 2nd Jun 2025 at 11:06
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master