Workflow Type: Galaxy
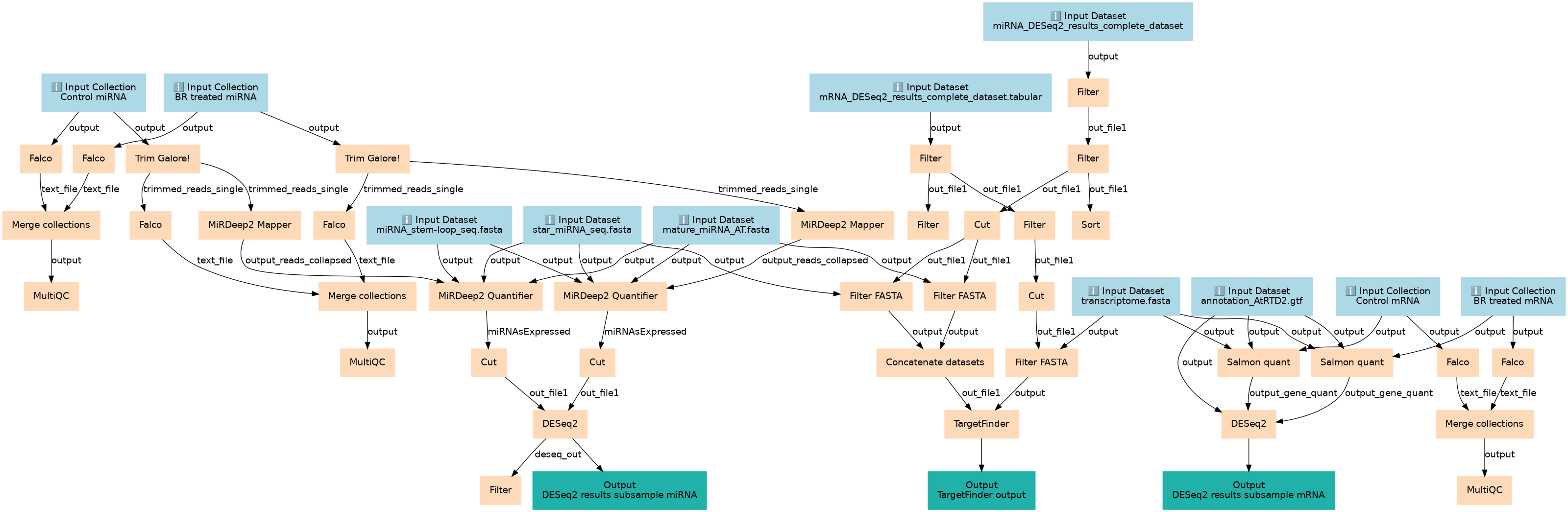
This workflow is generated from the GTN tutorial Whole transcriptome analysis of Arabidopsis thaliana (https://gxy.io/GTN:T00292).
Associated Tutorial
This workflows is part of the tutorial Whole transcriptome analysis of Arabidopsis thaliana, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Pavankumar Videm
Tutorial Author(s): Cristóbal Gallardo, Pavankumar Videm, Beatriz Serrano-Solano
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| BR treated mRNA | BR treated mRNA | Collection of BR treated mRNA FASTQs |
|
| BR treated miRNA | BR treated miRNA | Collection of BR treated miRNA FASTQs |
|
| Control mRNA | Control mRNA | Collection of control mRNA FASTQs |
|
| Control miRNA | Control miRNA | Collection of control miRNA FASTQs |
|
| annotation_AtRTD2.gtf | annotation_AtRTD2.gtf | Annotation in GTF format |
|
| mRNA_DESeq2_results_complete_dataset.tabular | mRNA_DESeq2_results_complete_dataset.tabular | DESeq2 results of complete mRNA dataset |
|
| mature_miRNA_AT.fasta | mature_miRNA_AT.fasta | mature miRNA sequences |
|
| miRNA_DESeq2_results_complete_dataset | miRNA_DESeq2_results_complete_dataset | DESeq2 results complete miRNA dataset |
|
| miRNA_stem-loop_seq.fasta | miRNA_stem-loop_seq.fasta | miRNA step loop sequences |
|
| star_miRNA_seq.fasta | star_miRNA_seq.fasta | miRNA star sequences |
|
| transcriptome.fasta | transcriptome.fasta | transcriptome sequences in FASTA format |
|
Steps
| ID | Name | Description |
|---|---|---|
| 11 | Falco | toolshed.g2.bx.psu.edu/repos/iuc/falco/falco/1.2.4+galaxy0 |
| 12 | Trim Galore! | toolshed.g2.bx.psu.edu/repos/bgruening/trim_galore/trim_galore/0.6.10+galaxy0 |
| 13 | Falco | toolshed.g2.bx.psu.edu/repos/iuc/falco/falco/1.2.4+galaxy0 |
| 14 | Trim Galore! | toolshed.g2.bx.psu.edu/repos/bgruening/trim_galore/trim_galore/0.6.10+galaxy0 |
| 15 | Filter | Filter1 |
| 16 | Filter | Filter1 |
| 17 | Falco | toolshed.g2.bx.psu.edu/repos/iuc/falco/falco/1.2.4+galaxy0 |
| 18 | Falco | toolshed.g2.bx.psu.edu/repos/iuc/falco/falco/1.2.4+galaxy0 |
| 19 | Salmon quant | toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/1.10.1+galaxy2 |
| 20 | Salmon quant | toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/1.10.1+galaxy2 |
| 21 | Falco | toolshed.g2.bx.psu.edu/repos/iuc/falco/falco/1.2.4+galaxy0 |
| 22 | MiRDeep2 Mapper | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_mapper/rbc_mirdeep2_mapper/2.0.0.8.1 |
| 23 | Merge collections | __MERGE_COLLECTION__ |
| 24 | Falco | toolshed.g2.bx.psu.edu/repos/iuc/falco/falco/1.2.4+galaxy0 |
| 25 | MiRDeep2 Mapper | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_mapper/rbc_mirdeep2_mapper/2.0.0.8.1 |
| 26 | Filter | Filter1 |
| 27 | Filter | Filter1 |
| 28 | Filter | Filter1 |
| 29 | Merge collections | __MERGE_COLLECTION__ |
| 30 | DESeq2 | toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.40.8+galaxy0 |
| 31 | MiRDeep2 Quantifier | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_quantifier/rbc_mirdeep2_quantifier/2.0.0 |
| 32 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.27+galaxy3 |
| 33 | Merge collections | __MERGE_COLLECTION__ |
| 34 | MiRDeep2 Quantifier | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_quantifier/rbc_mirdeep2_quantifier/2.0.0 |
| 35 | Sort | sort1 |
| 36 | Cut | Cut1 |
| 37 | Cut | Cut1 |
| 38 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.27+galaxy3 |
| 39 | Cut | Cut1 |
| 40 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.8+galaxy1 |
| 41 | Cut | Cut1 |
| 42 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.3 |
| 43 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.3 |
| 44 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.3 |
| 45 | DESeq2 | toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.40.8+galaxy0 |
| 46 | Concatenate datasets | cat1 |
| 47 | Filter | Filter1 |
| 48 | TargetFinder | toolshed.g2.bx.psu.edu/repos/rnateam/targetfinder/targetfinder/1.7.0+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| DESeq2 results subsample mRNA | DESeq2 results subsample mRNA | n/a |
|
| DESeq2 results subsample miRNA | DESeq2 results subsample miRNA | n/a |
|
| TargetFinder output | TargetFinder output | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 1633 Downloads: 287 Runs: 1
Created: 2nd Jun 2025 at 11:06
Last updated: 11th Aug 2025 at 14:14
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master 3.0
3.0