Workflow Type: Galaxy
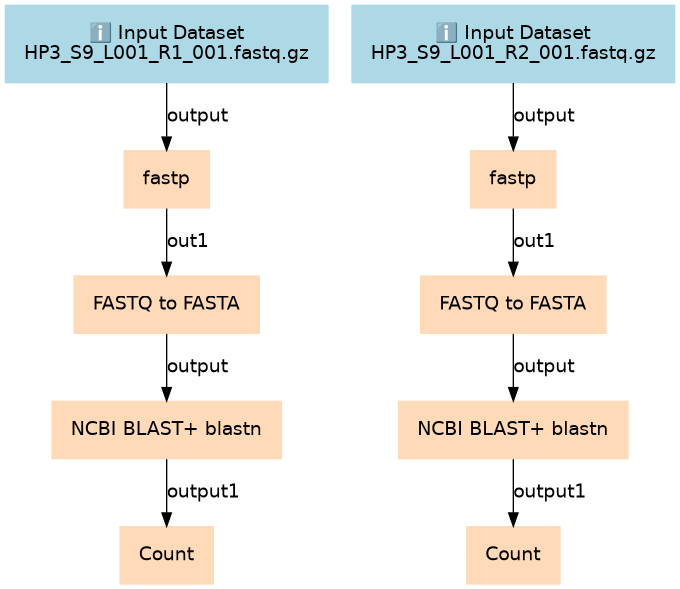
Final output is a table of Taxa and how many sequences aligned with them. This table can be used to then visualize via a pie chart.
Associated Tutorial
This workflows is part of the tutorial Taxonomic Analysis of eDNA, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): John Beliveau
Tutorial Author(s): John Beliveau
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| HP3_S9_L001_R1_001.fastq.gz | HP3_S9_L001_R1_001.fastq.gz | n/a |
|
| HP3_S9_L001_R2_001.fastq.gz | HP3_S9_L001_R2_001.fastq.gz | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.24.1+galaxy0 |
| 3 | fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.24.1+galaxy0 |
| 4 | FASTQ to FASTA | toolshed.g2.bx.psu.edu/repos/devteam/fastq_to_fasta/cshl_fastq_to_fasta/1.0.2+galaxy2 |
| 5 | FASTQ to FASTA | toolshed.g2.bx.psu.edu/repos/devteam/fastq_to_fasta/cshl_fastq_to_fasta/1.0.2+galaxy2 |
| 6 | NCBI BLAST+ blastn | toolshed.g2.bx.psu.edu/repos/devteam/ncbi_blast_plus/ncbi_blastn_wrapper/2.16.0+galaxy0 |
| 7 | NCBI BLAST+ blastn | toolshed.g2.bx.psu.edu/repos/devteam/ncbi_blast_plus/ncbi_blastn_wrapper/2.16.0+galaxy0 |
| 8 | Count | Count1 |
| 9 | Count | Count1 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 1420 Downloads: 196 Runs: 0
Created: 2nd Jun 2025 at 14:16
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master