Workflow Type: Galaxy
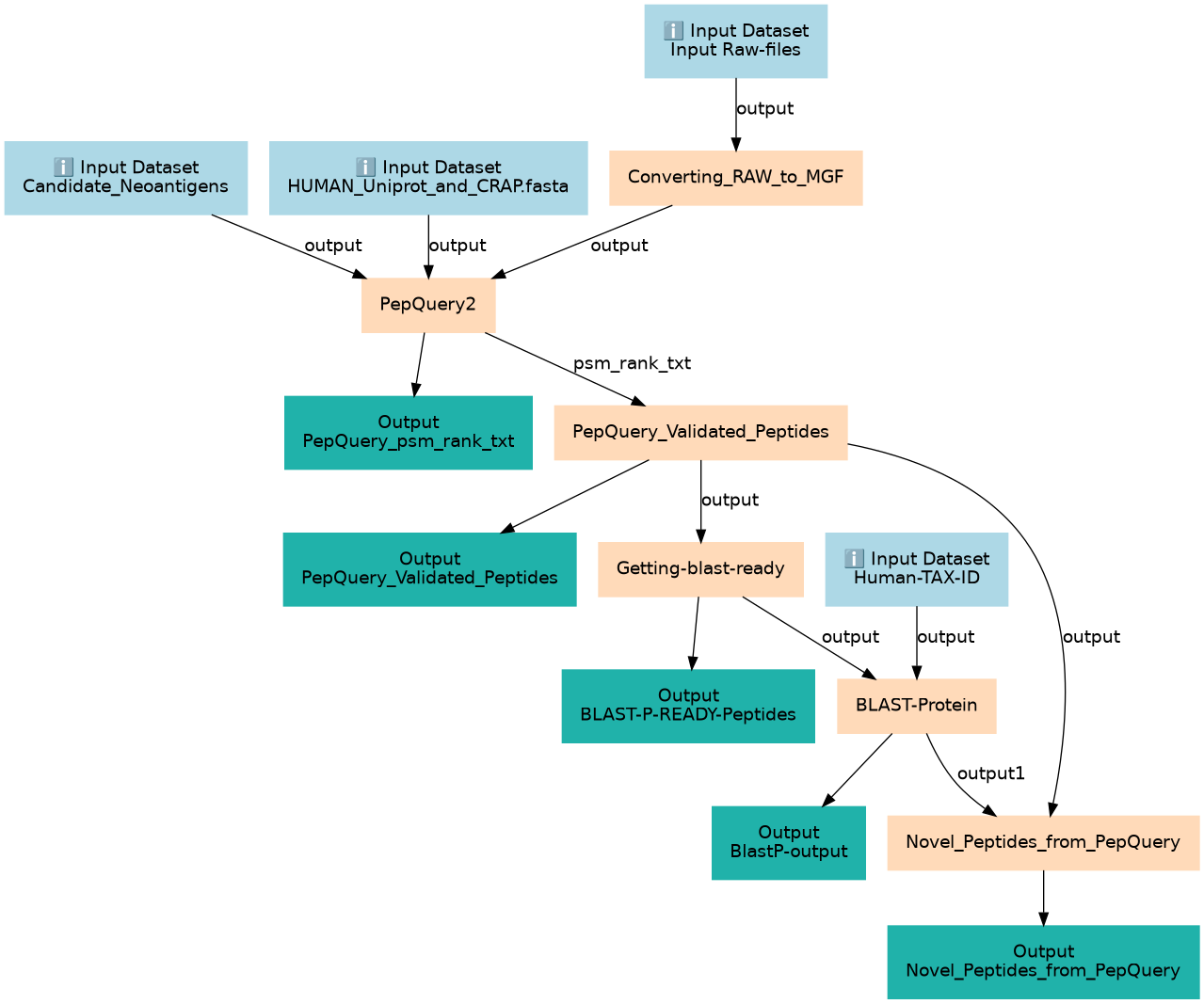
Validate the NeoAntigen Candidates from FragPipe discovery through the PepQuery Novel search
Associated Tutorial
This workflows is part of the tutorial Neoantigen 3: PepQuery2 Verification, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): GalaxyP
Tutorial Author(s): Subina Mehta, Katherine Do, James Johnson
Tutorial Contributor(s): Pratik Jagtap, Timothy J. Griffin, Subina Mehta, Saskia Hiltemann, Katherine Do
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Candidate_Neoantigens | #main/Candidate_Neoantigens | Candidate Neoantigens from Fragpipe Workflow |
|
| HUMAN_Uniprot_and_CRAP.fasta | #main/HUMAN_Uniprot_and_CRAP.fasta | Known human and contaminant protein fasta database |
|
| Human-TAX-ID | #main/Human-TAX-ID | this is for blastP taxid restrictions- homo sapiens-9606 |
|
| Input Raw-files | #main/Input Raw-files | RAW File |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Converting_RAW_to_MGF | Converting Raw to MGF toolshed.g2.bx.psu.edu/repos/galaxyp/msconvert/msconvert/3.0.20287.2 |
| 5 | PepQuery2 | PepQuery against HUMAN and contaminants to know filter peptides that doesn't match to anything in the known Uniprot database toolshed.g2.bx.psu.edu/repos/galaxyp/pepquery2/pepquery2/2.0.2+galaxy2 |
| 6 | PepQuery_Validated_Peptides | Extract Peptides with the confident column as "Yes" toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 7 | Getting-blast-ready | toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 8 | BLAST-Protein | BLAST-Protein against swissprot, ref seq and NCBI NR and filtering for HUMAN only results toolshed.g2.bx.psu.edu/repos/devteam/ncbi_blast_plus/ncbi_blastp_wrapper/2.14.1+galaxy2 |
| 9 | Novel_Peptides_from_PepQuery | Extracting Novel peptides after BlastP toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| BLAST-P-READY-Peptides | #main/BLAST-P-READY-Peptides | n/a |
|
| BlastP-output | #main/BlastP-output | n/a |
|
| Novel_Peptides_from_PepQuery | #main/Novel_Peptides_from_PepQuery | n/a |
|
| PepQuery_Validated_Peptides | #main/PepQuery_Validated_Peptides | n/a |
|
| PepQuery_psm_rank_txt | #main/PepQuery_psm_rank_txt | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 988 Downloads: 157 Runs: 0
Created: 7th Jul 2025 at 14:15
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master