Workflow Type: Galaxy
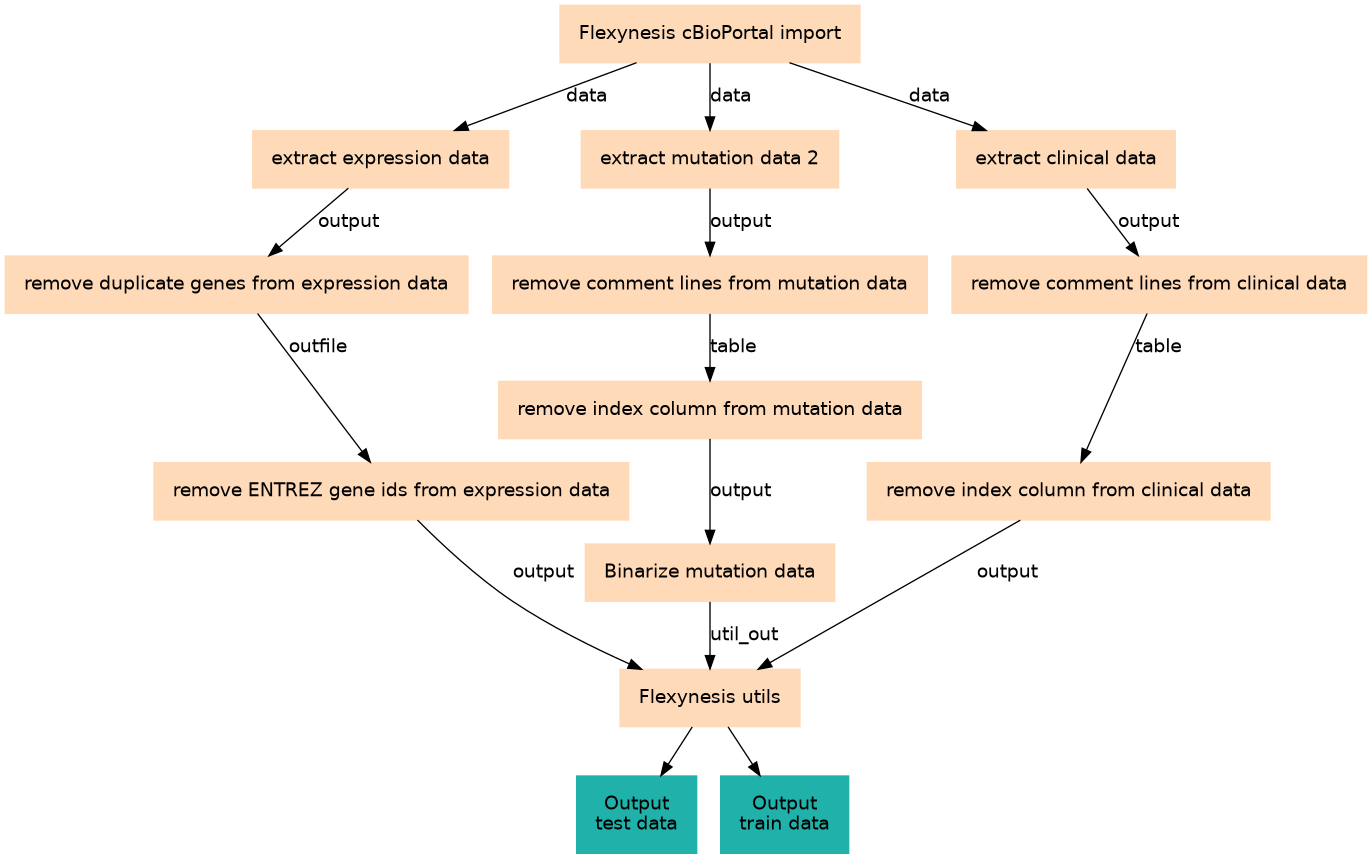
Download data from cBioPortal and prepare it for Flexynesis
Associated Tutorial
This workflows is part of the tutorial Prepare data from CbioPortal for Flexynesis integration, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Amirhossein Naghsh Nilchi
Tutorial Author(s): Amirhossein Naghsh Nilchi, Polina Polunina, Björn Grüning
Steps
| ID | Name | Description |
|---|---|---|
| 0 | Flexynesis cBioPortal import | toolshed.g2.bx.psu.edu/repos/bgruening/flexynesis_cbioportal_import/flexynesis_cbioportal_import/0.2.20+galaxy1 |
| 1 | extract expression data | __EXTRACT_DATASET__ |
| 2 | extract mutation data 2 | __EXTRACT_DATASET__ |
| 3 | extract clinical data | __EXTRACT_DATASET__ |
| 4 | remove duplicate genes from expression data | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.5+galaxy2 |
| 5 | remove comment lines from mutation data | toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy2 |
| 6 | remove comment lines from clinical data | toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy2 |
| 7 | remove ENTREZ gene ids from expression data | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.5+galaxy2 |
| 8 | remove index column from mutation data | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.5+galaxy2 |
| 9 | remove index column from clinical data | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.5+galaxy2 |
| 10 | Binarize mutation data | toolshed.g2.bx.psu.edu/repos/bgruening/flexynesis_utils/flexynesis_utils/0.2.20+galaxy3 |
| 11 | Flexynesis utils | toolshed.g2.bx.psu.edu/repos/bgruening/flexynesis_utils/flexynesis_utils/0.2.20+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| test data | test data | n/a |
|
| train data | train data | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 839 Downloads: 147 Runs: 0
Created: 11th Aug 2025 at 14:15
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master