Workflow Type: Galaxy
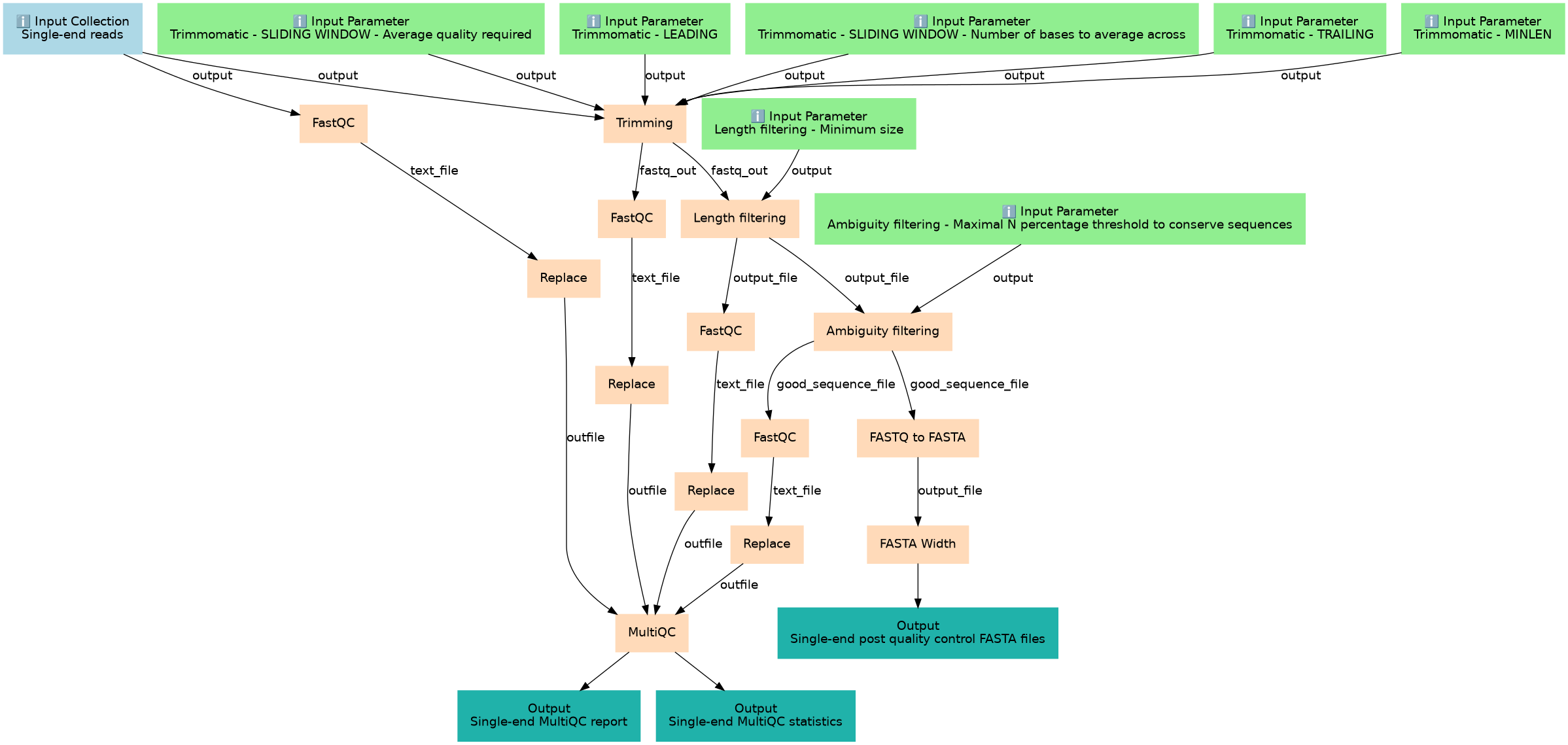
Quality control subworkflow for single-end reads.
Associated Tutorial
This workflows is part of the tutorial MGnify v5.0 Amplicon Pipeline, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): MGnify - EMBL, Rand Zoabi, Paul Zierep
Tutorial Author(s): Rand Zoabi
Tutorial Contributor(s): Paul Zierep, Saskia Hiltemann
Funder(s): German Competence Center Cloud Technologies for Data Management and Processing (de.KCD)
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Ambiguity filtering - Maximal N percentage threshold to conserve sequences | #main/Ambiguity filtering - Maximal N percentage threshold to conserve sequences | Maximal N percentage threshold to conserve sequences. |
|
| Length filtering - Minimum size | #main/Length filtering - Minimum size | Minimum sequence length. |
|
| Single-end reads | #main/Single-end reads | Single-end reads collection. |
|
| Trimmomatic - LEADING | #main/Trimmomatic - LEADING | Cut bases off the start of a read, if below a threshold quality. |
|
| Trimmomatic - MINLEN | #main/Trimmomatic - MINLEN | Minimum length of reads to be kept. |
|
| Trimmomatic - SLIDING WINDOW - Average quality required | #main/Trimmomatic - SLIDING WINDOW - Average quality required | Average quality required. |
|
| Trimmomatic - SLIDING WINDOW - Number of bases to average across | #main/Trimmomatic - SLIDING WINDOW - Number of bases to average across | Number of bases to average across. |
|
| Trimmomatic - TRAILING | #main/Trimmomatic - TRAILING | Cut bases off the end of a read, if below a threshold quality. |
|
Steps
| ID | Name | Description |
|---|---|---|
| 8 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 9 | Trimming | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.39+galaxy2 |
| 10 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/9.5+galaxy0 |
| 11 | Length filtering | toolshed.g2.bx.psu.edu/repos/devteam/fastq_filter/fastq_filter/1.1.5+galaxy2 |
| 12 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 13 | Ambiguity filtering | toolshed.g2.bx.psu.edu/repos/iuc/prinseq/prinseq/0.20.4+galaxy2 |
| 14 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 15 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/9.5+galaxy0 |
| 16 | FASTQ to FASTA | toolshed.g2.bx.psu.edu/repos/devteam/fastqtofasta/fastq_to_fasta_python/1.1.5+galaxy2 |
| 17 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 18 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/9.5+galaxy0 |
| 19 | FASTA Width | toolshed.g2.bx.psu.edu/repos/devteam/fasta_formatter/cshl_fasta_formatter/1.0.1+galaxy2 |
| 20 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/9.5+galaxy0 |
| 21 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.27+galaxy3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Single-end MultiQC report | #main/Single-end MultiQC report | n/a |
|
| Single-end MultiQC statistics | #main/Single-end MultiQC statistics | n/a |
|
| Single-end post quality control FASTA files | #main/Single-end post quality control FASTA files | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 869 Downloads: 143 Runs: 0
Created: 11th Aug 2025 at 14:15
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master