Workflow Type: Galaxy
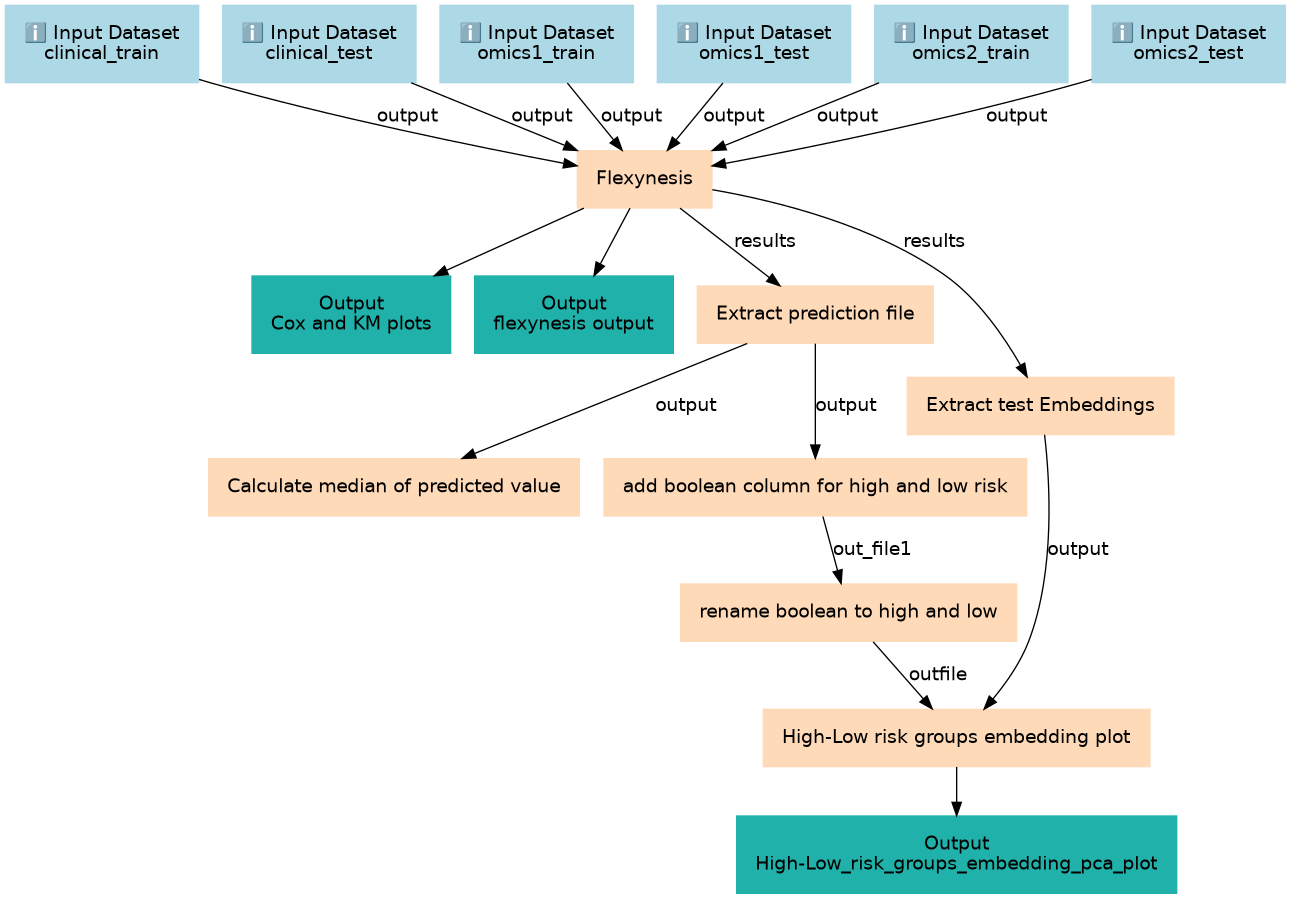
This workflow shows Flexynesis usage for predicting survival markers from a multi-omics dataset
Associated Tutorial
This workflows is part of the tutorial Identifing Survival Markers of Brain tumor with Flexynesis, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Amirhossein Naghsh Nilchi
Tutorial Author(s): Amirhossein Naghsh Nilchi, Björn Grüning
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| clinical_test | clinical_test | clinical test file |
|
| clinical_train | clinical_train | clinical training file |
|
| omics1_test | omics1_test | omics 1 test file |
|
| omics1_train | omics1_train | omics 1 training file |
|
| omics2_test | omics2_test | omics 2 test file |
|
| omics2_train | omics2_train | omics 2 training file |
|
Steps
| ID | Name | Description |
|---|---|---|
| 6 | Flexynesis | toolshed.g2.bx.psu.edu/repos/bgruening/flexynesis/flexynesis/0.2.20+galaxy3 |
| 7 | Extract prediction file | __EXTRACT_DATASET__ |
| 8 | Extract test Embeddings | __EXTRACT_DATASET__ |
| 9 | Calculate median of predicted value | toolshed.g2.bx.psu.edu/repos/iuc/datamash_ops/datamash_ops/1.9+galaxy0 |
| 10 | add boolean column for high and low risk | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.1 |
| 11 | rename boolean to high and low | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.5+galaxy2 |
| 12 | High-Low risk groups embedding plot | toolshed.g2.bx.psu.edu/repos/bgruening/flexynesis_plot/flexynesis_plot/0.2.20+galaxy3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Cox and KM plots | Cox and KM plots | n/a |
|
| flexynesis output | flexynesis output | n/a |
|
| High-Low_risk_groups_embedding_pca_plot | High-Low_risk_groups_embedding_pca_plot | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 767 Downloads: 123 Runs: 0
Created: 18th Aug 2025 at 14:16
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master